Abstract
We conducted a cytogenetic study of sexual lines of Boechera stricta and Boechera holboellii (2n = 14) and seven diploid apomictic accessions of their interspecific hybrid Boechera divaricarpa and B. holboellii (2n = 14 or 15). By studying chromosome morphology, rDNA repeats, genome painting, male meiosis, pollen morphology, and flow-cytometry seed screens, we revealed an unexpected plethora of chromosome forms, pairing behavior, and hybrid composition in all apomictic lines. Genome painting demonstrated that the apomicts are alloploid with variable numbers of B. stricta and B. holboellii-like chromosomes. We assume that large-scale homeologous chromosome substitutions took place in the apomictic hybrids that resulted from recurrent diploid–polyploid transitions through restitutional meiosis and polyploidy–diploid transitions through reductional meiosis. A second peculiarity was the presence of a largely heterochromatic chromosome (Het) in all apomictic accessions (2n = 14 and 15) and an additional smaller chromosome (Del) in the aneuploids (2n = 15). Both chromosomes share repetitive pericentromere repeats with those from the sexual B. stricta, suggesting that they originated from this species. Pairing and behavior at meiosis I of the Het share features with both Y and B chromosomes and suggest that the Del arose from a translocation event or homeologous recombination between a B. holboellii (or related taxon) and a B. stricta chromosome. Based on its presence exclusively in apomictic accessions, we propose that the Het chromosome plays a role in the genetic control of apomixis.
Keywords: apomixis, chromosome evolution, genome painting, heterochromatic chromosome, homeologous substitution
The genus Boechera (formerly Arabis), Böcher's rock cress, a biennial or perennial genus of the Brassicaceae, is distributed from Alaska through a greater part of North America to Greenland. The genus is monophyletic, has a basic chromosome number of x = 7 (1), and has diploid sexuals, as well as diploid, aneuploid, and polyploid apomictics that reproduce asexually through seeds. Classification of Boechera is mostly based on trichome and silique morphology (2, 3). The genus comprises 50–80 species whose remarkably high levels of polymorphism are linked with polyploidy, aneuploidy, and interspecific hybridization (4). This variation, along with its widespread distribution in pristine habitats, diverse modes of reproduction, hybridization, and close relationship to Arabidopsis thaliana, makes Boech era a promising model for studies on speciation, apomixis, taxonomy, evolution, and phylogeography (5).
The Boechera holboellii complex includes B. holboellii, Boechera stricta (Arabis drummondii), and the hybrid Boechera divaricarpa. The three species have traditionally been delimited based on trichome and silique morphology, with B. divaricarpa having an intermediate phenotype. Morphological and cpDNA studies (3) have demonstrated that B. stricta is monophyletic, whereas elevated chloroplast diversity demonstrates that B. holboellii is polyphyletic. Its sequence and microsatellite analyses have shown that B. divaricarpa arose through hybridization between sexual B. stricta and B. holboellii or a closely related species (3, 5, 6). The level of allelic variation is comparable between B. divaricarpa and B. holboellii, and a low number of species-specific alleles suggests that the hybrid originated recently (6). Multiple evolutionary origins of triploidy in Boechera imply that the apomictic phenotype also has repeatedly been expressed from a diploid sexual background (7, 8).
Chromosome numbers in apomictic B. holboellii can vary from 2n = 14, 15, 21, 22, 28, 35, up to 42. Most B. stricta accessions are sexual diploids (2n = 14), although a small number of triploid apomicts have been reported (5, 7, 8). In his study of male and female meiosis of diploid and triploid B. holboellii from Greenland and Alaska, Böcher (9) identified apomeiosis and other meiotic variants, including abnormal pairing and skewed chromosome segregations, phenomena that may imply karyotype heteromorphy. A second intriguing cytogenetic aspect of apomictic Boechera lineages is the geographically widespread occurrence of supernumerary chromosomes (9), which were later identified as B chromosomes by using flow-cytometric, karyological, and DNA sequence analyses (7, 8, 10).
We selected a number of diploid sexual and apomictic accessions for detailed karyotype analyses of DAPI-stained chromosome complements in combination with rDNA probe FISH. The same material also was used for studying meiotic pairing and chiasma formation, chromosome segregation, and seed formation by using the flow-cytometric seed screen (FCSS) (11). Inspired by the karyotype heteromorphy and the lack of chromosome pairing in apomictic B. divaricarpa and some B. holboellii accessions, we applied one- and two-color genomic in situ hybridization (GISH) to study their putative hybrid nature. Although polyploidy is widespread in this complex, we only focused on diploids and aneuploids to simplify chromosome identification and facilitate interpretations of meiotic aberrations. Thus, our data elucidate only part of the potential chromosome diversity of the B. holboellii complex, which encompasses diverse sexual and apomictic taxa of varying ploidy (6).
Results
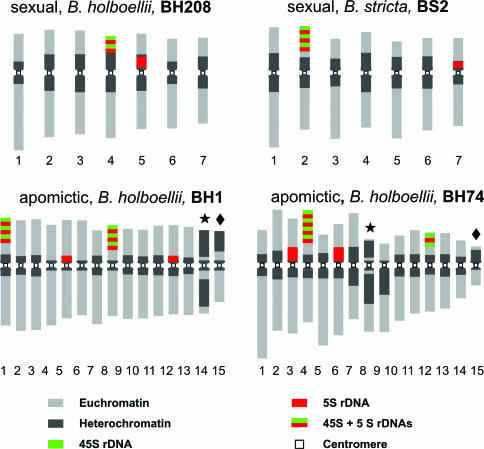
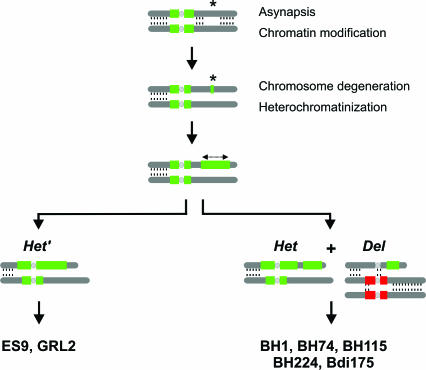
The karyotypes of the sexual B. holboellii and B. stricta display metacentric or subtelocentric chromosomes that measure 1.2–3.5 μm and 1.5–5.8 μm, respectively. They have one nucleolar organizer region pair with overlapping 45S and 5S rDNA and one additional pair with only 5S rDNA (Fig. 1). In contrast, the karyotypes of the seven apomictic accessions were strikingly dissimilar, and chromosomes could not be matched to form putative homologues (Fig. 1). Part of this variation was further substantiated by rDNA FISH, which clearly demonstrated repeat-size polymorphism, especially in the aneuploid BH74 apomict. Apomicts also displayed a variable number of chromosomes with brightly fluorescing pericentromere bands, the most notable of which included one with large blocks of heterochromatin, hereafter referred to as Het, and one small (deletion) chromosome, referred to as Del (Fig. 1). The 2n = 14 apomicts ES9 (B. divaricarpa) and GRL2 (B. holboellii) had only the Het chromosomes, but the heterochromatin block is larger than that of the Het in the 15-chromosome apomicts.
Fig. 1.
Ideograms of the sexual diploid B. holboellii (BH208) and B. stricta (BS2) and the 15-chromosome B. holboellii apomicts (BH1 and BH74) based on DAPI fluorescence and FISH with the 45S and 5S rDNA probes. The chromosome sets of the aneuploid BH1 and BH74 demonstrate excessive chromosomal heteromorphy and the two aberrant chromosomes, Het (star) and Del (diamond).
In the sexual accessions, meiosis was regular as expected, with fully paired chromosomes at pachytene and seven bivalents at metaphase I, each containing one to three chiasmata. Heterochromatin differentiation was most obvious at pachytene, resembling the typical Arabidopsis pachytene pattern of heterochromatin only around centromeres and nucleolar organizer regions. Pericentromere heterochromatin of B. stricta was two to three times longer than that of B. holboellii, which is concordant with previously described metaphase length differences and ≈8% total genomic DNA size difference between the species (R. Vašut, personal communication) (7, 8). Pollen was regularly shaped, ≈100% viable, and measured 12.3 μm on average. Our sample of B. stricta also showed some unreduced pollen. FCSS revealed the expected peaks for 2C (embryo) and 3C (endosperm).
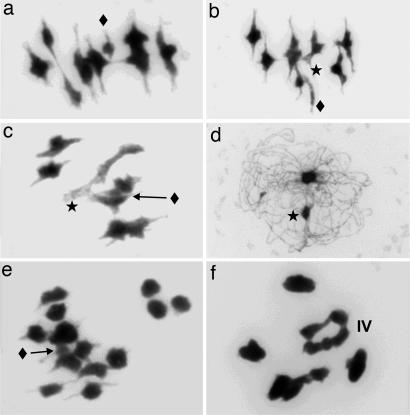
Apomictic Boecheras are facultative, and thus they can produce seed by both apomictic and sexual mechanisms. The production of apomictic versus sexual seed displays quantitative variation between accessions (12). For the purpose of this article, we labeled accessions that produced any proportion of seed apomictically as “apomictic” (Table 1). The apomicts display striking variability with respect to chromosome pairing, chiasma formation, and chromosome transmission in male meiosis. The synaptic apomicts BH1, BH115, BH224, and ES9 had normal pachytene and regular chiasmata. The Het was easily identified at metaphase I as a Y-like, heteropycnotic chromosome associated with one other chromosome (Fig. 2 b and c), whereas the Del remained unpaired (Fig. 2a) or associated with two larger chromosomes to form a heteromorphic trivalent (Fig. 2 b and c). BH115 also displayed a conspicuous quadrivalent (Fig. 2f), which we interpret as heterozygosity for a reciprocal translocation.
Table 1.
Overview of all Boechera species and accessions, with mode of reproduction (MOR), chromosome number (2n =), origin/place of collection, characteristics of male meiosis, pollen size distribution, FCSS, and GISH showing the number of chromosomes with the B. holboellii-specific (H) and the B. stricta-specific (S) repeats
| Accession no. | Morphology | MOR | 2n = | Origin/place of collection | Aberrant chromosome | Meiosis | Pollen size | FCSS | GISH |
|
|---|---|---|---|---|---|---|---|---|---|---|
| H | S | |||||||||
| BH208 | B. holboellii | Sex | 14 | Charles Gulch, Ravalli Co., MT | — | Synaptic, reductional | n | 2C + 3C | 14 | 0 |
| BS2 | B. stricta | Sex | 14 | Ohio City, Gunnison Co., CO | — | Synaptic, reductional | n and few 2n | 2C + 3C | 0 | 14 |
| ES6 | B. stricta | Sex | 14 | Taylor River, Gunnison Co., CO | — | Synaptic, reductional | n | 2C + 3C | 0 | 14 |
| ES9 | B. divaricarpa | Apo | 14 | Vipond Park, Beaverhead Co., MT | Het′ | Synaptic, restitutional | n–2n/irregular | 2C + 6C | 7 | 7 |
| GRL2 | B. holboellii | Apo | 14 | Greenland; Botanical Garden, Copenhagen, Denmark | Het′ | — | — | — | 9 | 5 |
| BH1 | B. holboellii | Apo | 15 | Wallowa Mountains, OR | Het + Del | Synaptic, reductional | n | 2C + 5C | 11 | 4 |
| BH74 | B. holboellii | Apo | 15 | Ranch Creek, Granite Co., MT | Het + Del | Asynaptic, restitutional | n–2n/irregular | 2C + 6C | 4 | 11 |
| BH115 | B. holboellii | Apo | 15 | Birch Creek, Ravalli Co., MT | Het + Del | Synaptic, reductional and restitutional + translocation | Mostly 2n | 2C + 6C | 5 | 10 |
| BH224 | B. holboellii | Apo | 15 | Bandy Ranch, Missoula Co., MT | Het + Del | Synaptic, reductional and restitutional | n–2n/irregular | 2C + 6C | 11 | 4 |
| Bdi175 | B. divaricarpa | Apo | 15 | Lower Storm Lake, Deerlodge Co., MT | Het + Del | Asynaptic, restitutional | 2n/irregular | 2C + 6C | 9 | 6 |
The collectors include T.M.-O. (BH208, BH1, BH74, BH115, and Bdi175), B. A. Roy (BS2 and ES6), J. McKay (ES9), K. Boutilier (GRL2), and M. Marler (BH224). Co., county; —, not applicable.
Fig. 2.
Meiosis in DAPI-stained pollen mother cells of B. holboellii apomicts. The Het chromosome (star) is often clearly heteropycnotic and can form a heteromorphic bivalent. The Del chromosome (diamond) is much shorter than the other chromosomes and can associate with two other chromosomes to form a heteromorphic trivalent. (a–c) BH1 at metaphase I. (d and e) BH74 at asynaptic pachytene (d) and metaphase I (e). (f) BH115 at metaphase I.
Pollen in BH1 is comparable to that of the sexual diploid species and was inferred to be haploid resulting from reductional meiosis. Pollen size in ES9, BH74, and BH224 was n–2n in size, reflecting reductional or restitutional meiosis or aneuploidy, whereas Bdi175 was mostly 2n. As is evidenced by the FCSS analysis, female meiosis in BH1 was apomeiotic, whereas male meiosis was normal and reductional (Table 1). BH115 and BH224 were partly restitutional and formed first-division restitution gametes. BH74 and Bdi175 (Fig. 2 d and e) displayed severely disturbed pairing at pachytene and mostly univalents at metaphase I. Interestingly, the FCSS of BH74 demonstrated that only the larger (unreduced) pollen led to successful fertilization. The asynaptic meiosis and the hybrid morphology of Bdi175 suggested that at least some members of Boechera can be alloploid, which inspired us to conduct genome-painting experiments.
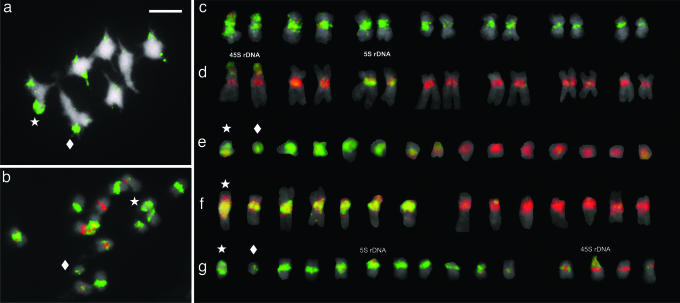
Using FISH with total genomic DNA from B. stricta as a probe and blocking with B. holboellii genomic DNA, we observed fluorescent foci only on the pericentromere regions of the B. stricta chromosomes. This finding was further confirmed in a two-color genome painting (Fig. 3 a–e), whose resultant species-specific chromosomal fluorescence patterns demonstrate that B. holboellii and B. stricta have undergone dramatic evolutionary changes in their pericentromere-repetitive sequences, and that the number of parental chromosomes can differ in each apomictic accession (Table 1). For example, apomictic accessions morphologically classified as B. divaricarpa had six B. stricta and nine B. holboellii chromosomes (BDi175) or seven B. stricta and seven B. holboellii chromosomes (ES9). Apomictic accessions that have the B. holboellii phenotype were highly variable, including 5 B. stricta and 9 B. holboellii chromosomes (GRL2), 4 B. stricta and 11 B. holboellii chromosomes (BH1), and 11 B. stricta and 4 B. holboellii chromosomes (BH74).
Fig. 3.
Genome painting of metaphase complements obtained according to the one- and two-color GISH. (a) One-color painting of a metaphase I complement of BH1. The green Het (stars) and Del (diamonds) indicate that these chromosomes have the B. stricta pericentromere repeat. (Scale bar: 3 μm.) (b) Two-color painting of a BH74 mitotic metaphase. (c–g) Karyograms of the two-color painting of the diploid sexual species B. stricta (c), B. holboellii (d), the B. divaricarpa apomicts BDi175 (e) and ES9 (f), and the B. holboellii apomict BH1 (g).Chromosomes with green signals have the B. stricta repeat. The red signals denote to the B. holboellii origin. The yellow signals in the chromosomes sets of d and g resulted from rDNA hybridizations from genomic DNAs from both B. stricta and B. holboellii genomic DNA.
Discussion
This study on chromosome morphology, meiosis, and mode of reproduction of diploid (2n = 14) and aneuploid (2n = 15) members of the B. holboellii complex has revealed extensive chromosomal variation in apomictic lines, which is concordant with previous studies on genome plasticity in this genus (4, 7, 9). Not only have we shown large differences among apomictic karyotypes (Fig. 1), disturbed meiosis (Fig. 2), variable reproduction, and the presence of aberrant chromosomes (Figs. 1–3 and Table 1), the results of our genome painting also show that the apomicts are interspecific hybrids but differ in the numbers of parental chromosomes (Fig. 3 and Table 1). The aberrant chromosomes, previously referred to as B chromosomes (8, 10), were either heterochromatic or very small. Using better microscopy and cytological techniques, we show here that there are two different aberrant chromosomes in the 2n = 15 apomicts, Het and Del.
How can one explain the pattern of GISH karyotypes that we have observed in the apomictic accessions studied here? To begin with, it is not surprising that the two sexual B. stricta accessions demonstrate species-specific pericentromere-repetitive DNA because this species is monophyletic (6). Similarly, all chromosomes of the sexual B. holboellii line shared a set of B. holbellii-like-specific, pericentromere-repetitive DNA. Furthermore, although the GISH karyotypes of the apomictic accessions suggest a mixture of both B. stricta and B. holboellii chromosomes, it is unclear whether the repeat DNA responsible for the B. holboellii GISH patterns are truly specific to B. holboellii or some other closely related species. For example, it is hypothesized that B. divaricarpa is the hybrid between B. stricta and B. holboellii (3, 6), although there is growing evidence that B. holboellii is not one of the parents in the hybridization event, as is evidenced by molecular genetics (3, 5, 6) and crossing data (13). Our results further demonstrate the taxonomical difficulties in delimiting species because of incongruities between phenotype and karyotype, suggesting that traditional taxonomical traits may be controlled by only a few genes of large effect. Regardless, it is clear that the apomictic genomes analyzed here are both hybrid in origin and characterized by Het and Del chromosomes.
The only seven B. stricta and seven B. holboellii-like chromosome apomict is ES9, reflecting a recently formed hybrid between the two parental species. Alternatively, deviations from this 1:1 ratio in the other apomictic accessions, including an excess of either parental chromosome type, point to a complex pattern of hybrid origin and gene flow. A number of diploid Boechera species have apomeiotic potential (13), as is supported by seed and pollen analyses (12) and the few unreduced pollen that we found in the pollen sample of B. stricta. Many of these diploids are sexual, and hence it appears that apomixis per se is not a prerequisite source of unreduced gametes. Laboratory crosses (13) demonstrated that triploidy could be generated in a number of ways, including through crosses between diploids (fusion of unreduced and reduced gametes) or between diploids and tetraploids (fusion of reduced gametes). In addition, diploid B. stricta, B. holboellii, and B. divaricarpa all have the potential to produce unreduced gametes (13), and it appears that at least some diploid and triploid apomicts can either fertilize or be fertilized by other accessions or both (13). Finally, in line with De Wet's cycle (14), it is plausible that tetraploids can parthenogenically form diploid offspring with unusual complements of chromosomes.
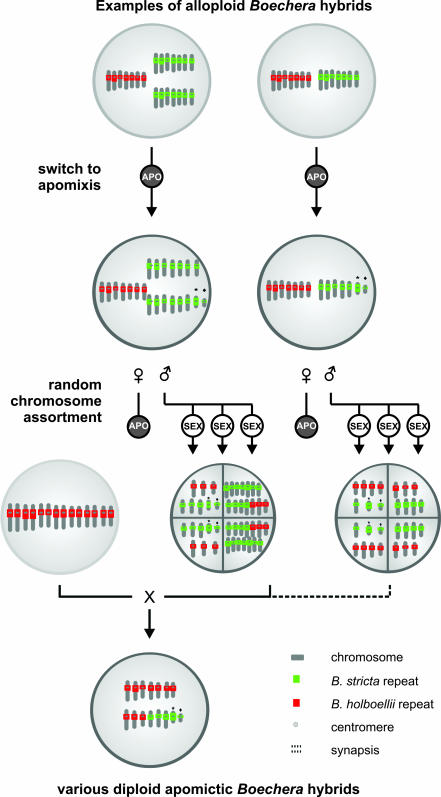
There are thus multiple pathways that could explain how a diploid hybrid gives rise to diploid apomictic karyotypes with different numbers of parental chromosomes. Upon the origin of an F1 diploid hybrid, meiotic chromosome segregation (with or without synapsis) and reduction would lead to gametes characterized by differing numbers of parental homologues (Fig. 4). Syngamy, with reduced or unreduced gametes from either pure or hybrid individuals, followed by further meioses could thus generate the variation in parental chromosome ratios documented here. Because all apomicts have at least four B. stricta chromosomes, including the two aberrant Het and Del chromosomes, it is conceivable that a specific chromosome combination is more favorable or even essential for the production of viable apomictic offspring. Furthermore, it has been suggested that the relative expression of B. stricta- and B. holboellii-specific alleles may explain how hybrid individuals come to exhibit the diagnostic silique and trichome phenotypes of one of their sexual parents (8). However, it is unclear why the BH74 apomictic accession exhibited the B. holboellii phenotype regardless of its genome being mostly B. stricta.
Fig. 4.
Model for explaining chromosome substitutions in Boechera. Several processes may lead to allopolyploid hybrids, but only two examples are drawn here: an allodiploid resulting from 2n gametes and an allotriploid resulting from a reduced B. holboellii gamete and an unreduced (2n) gamete from B. stricta that may come from B. holboellii as well. Once such a hybrid (either amphitriploid or amphidiploid) adopts the apomictic nature, it can donate various combinations of red and green chromosomes through random interchromosomal recombination in the male meiosis.
It appears that numerous apomictic lineages (i.e., clones) have been generated repeatedly from different sexual ancestors within this complex (7, 8, 10). The presence of diploid apomictic lineages in the B. holboellii complex additionally shows that polyploidy is not a requirement for apomixis expression. Because apomeiotic egg formation appears to be an evolutionary old trait that can be expressed in sexual members of the genus, we postulate that hybridization and its associated genomic stress (15–19) could rapidly induce the gene-expression changes required to switch to the apomictic pathway. Mechanisms that can alter gene expression profiles in newly formed allopolyploids include aberrant or novel interactions of diverged transcriptional regulators (18, 20). It also is clear that hybrid transcriptomes demonstrate a range of gene action changes relative to the parental genomes (21). The hybrid genomes of different Boechera accessions are expected to have been affected in a similar fashion to give rise to the apomixis switch, although it is unclear how.
We next examined the origin and nature of the aberrant chromosomes. The consistent presence of the aberrant chromosomes in the apomictic clones suggests a key role in controlling the apomixis trait, but recent genetic analysis suggests that the Het chromosome does not carry a dominant apomixis gene (22). Intriguingly, both hemizygosity and heteromorphism for an apospory-specific genomic region have similarly been identified in Pennisetum squamulatum and Cenchrus ciliaris (23–25). The work undertaken here does not shed light on the action of the Het and Del chromosomes with respect to apomixis expression, and thus we focus on the origin and evolution of the elements.
The Het chromosomes are characterized by a large B. stricta repeat block (Fig. 5), which, together with DNA sequence data (10), suggests that they originated from B. stricta. Metaphase I and genome painting of the BH1, BH115, and BH224 also shows that the Het can form heteromorphic chromosome associations with a B. stricta chromosome, whereas the Del associates through chiasmate bonds with a B. holboellii bivalent, a remarkable difference considering that both chromosomes have a B. stricta-specific repeat. The conspicuous B. stricta repeat region and the heteropycnotic behavior of the Het chromosome suggest large-scale repeat amplification and heterochromatin formation, apparently accompanied with an extension of the unpaired region. The Del chromosome, with its B. stricta pericentromere region and pairing capability with a B. holboellii pair, is likely a translocation chromosome formed from a proximal B. stricta and a distal B. holboellii segment, respectively. Although it is unclear whether the Het and Del chromosomes have a common origin, their structure and meiotic dynamics demonstrate that they are evolving independently of one another. The recent publication of a genetic linkage map for B. stricta, and information about syntenic relationships to Arabidopsis, should provide the opportunity to further study the origin of the Het and Del chromosomes (26).
Fig. 5.
Schematic representation of the possible origin of aberrant Het′ and Het + Del chromosomes in the 14- and 15-chromosome apomictic Boechera accessions. We assume asynapsis as a possible switch to modify chromatin and epigenetic status of a chromosome region. This region is then involved in a stepwise chromosome degeneration and heterochromatinization, leading ultimately to the Het chromosome. The Del chromosome is considered a translocation or recombinant chromosome between a B. stricta and B. holboellii chromosome.
The heteromorphic behavior of the Het chromosome strongly suggests failure or impairment of homologous pairing of the ancestral bivalent (Fig. 5), a widely accepted view on the evolutionary origin of heteromorphic sex chromosomes. This first step could have been initiated by interspecific hybridization, which has been shown to affect species-specific chromosomal modifications and uniparental gene silencing in allopolyploids (16). Such modifications become constitutive and undergo stable inheritance in subsequent generations, thus leading to permanent change and gradual accumulation of heterochromatin (Fig. 5) reflective of Y chromosome evolution (27).
One of the most intriguing facts about the Het chromosome is its nature and function. The Het shares many characteristics of a group of heterochromatinized elements, including B chromosomes (28), Y chromosomes (29), and supernumerary fragments (30), all of which have highly compacted chromatin typically identified by using various chromosome-staining methods. Many, but not all, studies of such chromosomes have demonstrated high proportions of repetitive DNA, methylated DNA, and modified histones. It also is assumed that such chromosomes have little to no single copy sequences, and that they accumulate deleterious mutations through time (31).
The distinction between cases of such (partly) degenerated chromosomes for the most part is clear. B chromosomes are parasitic, often with different numbers in individuals of the same population; they do not possess genes essential to the organism; and they never recombine with any of the other chromosomes. Y chromosomes are associated with the X chromosome and are essential for sex determination, but their number and molecular organization differ enormously between plants and animals. Supernumerary fragments, like the apospory-containing region in P. squamulatum (23, 25), are large hemizygous regions. However, a number of studies have claimed cases of chromosomes having intermediate properties with respect to B and Y chromosomes or supernumerary fragments. In this light, the aberrant chromosomes in Boechera share properties of both B (extra, but not sex-determining) and Y chromosome concepts (pairing and recombination with one other chromosome). Both its putative association with the apomixis phenotype and the heterochromatic nature of the Het chromosome hint at similarities to the supernumerary chromosome fragment of Pennisetum, whereby an absence of meiotic recombination has led to repetitive sequence amplification and heterochromatinization. Heterochromatin initiation and spread are not random, but rather appear to be under the epigenetic control of boundary proteins and small RNAs (32, 33). The evolutionary pathways that have ultimately led to the various Het and Del chromosome forms seen here thus likely include both directed (e.g., heterochromatinization) and stochastic (e.g., transposition insertions, mutation accumulation, chromatin modifications, and large-scale chromosomal rearrangements) processes, although it is unknown to what extent each has acted in the independently evolving apomictic lineages.
Materials and Methods
Plant Material.
The diploid sexual species B. holboellii (BH208) and B. stricta (BS2), the apomictic hybrid B. divaricarpa (BDi175), and four apomictic B. holboellii accessions (BH1, BH74, BH115, and BH224) were obtained from T.M.-O. and T.F.S.; B. stricta accession ES6 and the apomictic B. divaricarpa ES9 were from M.E.S.; and the apomictic GRL2 came from K. Boutilier (Plant Research International, Wageningen, The Netherlands). All accessions except for GRL2, which was obtained from the Botanical Garden of Copenhagen, Denmark, were collected from natural populations in North America (Table 1).
Chromosome Preparations.
Chromosome spread preparations were obtained from actively growing root tips and meiotic cell complements as described (2, 34).
DAPI Staining and Karyotype Analysis.
Chromosomes were counterstained with DAPI (4′,6-diamidine-2-phenylindole) in VECTASHIELD Mounting Medium (Vector Laboratories, Burlingame, CA) and studied under a fluorescence microscope. We captured well spread cell complements; measured the chromosome arms, heterochromatic regions, and secondary constrictions (nucleolar organizer regions); and optimized contrast with Adobe Photoshop (Adobe Systems, Mountain View, CA).
FISH.
For standard FISH, we used probes for 45S (pTA71) and 5S (pTA794) rDNA. DNA was labeled with either biotin-16-dUTP or digoxiginin-11-dUTP by using the nick translation kit (Roche, Mannheim, Germany). FISH followed methods described previously (2, 22, 34). For genome painting, we used total genomic DNA of the diploid sexual B. holboellii (BH208), B. stricta (BS2), and A. thaliana (Col). Probe DNA was labeled with either digoxiginin-11-dUTP or biotin-16-dUTP by using the nick translation kit (Roche). The hybridization signals were detected with FITC-conjugated anti-dig antibodies and amplified with FITC-conjugated rabbit anti-sheep antibodies for the digoxigenin-labeled probe, and with Texas red avidin for the biotin-labeled probe, which was amplified with biotinylated anti-avidin and Texas red avidin. For the single-color genome painting, we used 100 ng of the genomic DNA probe and 10 μg of blocking DNA (of the other parental genome) in 40 μl of hybridization buffer. For the two-color system, the hybridization mixtures contained both B. holboellii and B. stricta probes and blocking DNA (1:100 total genomic DNA) of A. thaliana. The preparations were counterstained and mounted as described previously.
Pollen Staining and Measurements.
Pollen was squeezed out in a drop of lactophenol acid, fuchsin-staining solution (2). Size distribution was compared with pollen of the sexual species with regular meiosis. We used these results as references for comparison with pollen size measurements of the apomictic accessions, considering the mode of male meiosis (in the same inflorescence) and nuclear DNA amounts by flow cytometry of the offspring seeds. For the apomictic accessions, we only measured regular stained pollen.
FCSS.
Embryo and endosperm ploidy from dry seeds were analyzed by using a Partec Flow Cytometer PA (Partec, Münster, Germany) following the FCSS technique (11). DNA content was examined in single- and bulk-seed samples (15) for each accession. Briefly, seeds were crushed on dry fine-grade sandpaper and rinsed in 2 ml of the DAPI staining solution [5 mM MgCl2/0.1 M NaCl/2.5 mM sodium citrate/0.1 M Tris/10% Triton X-100/2.2 μM DAPI (pH 7.0)]. The suspensions were filtered through a 20-μm mesh nylon sieve and incubated in the dark at 4°C for 30 min before flow-cytometric analysis.
Acknowledgments
We thank Kim Boutilier for the Greenland accession, Peter van Dijk for discussions about the manuscript, Dr. Ihsan Al-Shehbaz and Christoph Dobeš for help with the taxonomy of the plant samples, Boudewijn van Veen for assistance with fluorescence microscopy and digital processing, Radim Vašut for carrying out flow cytometry of total genomic DNA sizes, Sonja Prodanovic for the help with the FCSS, and Marie Luise-Voigt for enlightening discussions about Boechera and helpful comments on manuscript preparation.
Abbreviations
- FCSS
flow-cytometric seed screen
- GISH
genomic in situ hybridization.
Footnotes
The authors declare no conflict of interest.
References
- 1.Koch M, Bishop J, Mitchell-Olds T. Plant Biol. 1999;1:529–537. [Google Scholar]
- 2.Kantana L. Wageningen, The Netherlands: Wageningen Univ; 2005. PhD thesis. [Google Scholar]
- 3.Dobeš C, Mitchell-Olds T, Koch M. Mol Ecol. 2004;13:349–370. doi: 10.1046/j.1365-294x.2003.02064.x. [DOI] [PubMed] [Google Scholar]
- 4.Dobeš C, Sharbel TF, Koch M. Syst Biodiversity. 2007;5:321–331. [Google Scholar]
- 5.Koch MA, Dobeš C, Mitchell-Olds T. Mol Biol Evol. 2003;20:338–350. doi: 10.1093/molbev/msg046. [DOI] [PubMed] [Google Scholar]
- 6.Dobeš C, Mitchell-Olds T, Koch M. Am J Bot. 2004;91:2087–2101. doi: 10.3732/ajb.91.12.2087. [DOI] [PubMed] [Google Scholar]
- 7.Sharbel TF, Mitchell-Olds T. Heredity. 2001;87:59–68. doi: 10.1046/j.1365-2540.2001.00908.x. [DOI] [PubMed] [Google Scholar]
- 8.Sharbel TF, Mitchell-Olds T, Dobeš C, Kantama L, de Jong H. Cytogenet Genome Res. 2005;109:283–292. doi: 10.1159/000082411. [DOI] [PubMed] [Google Scholar]
- 9.Böcher TW. Biol Skr Vid Selsk. 1951;6:1–59. [Google Scholar]
- 10.Sharbel TF, Voigt M-L, Mitchell-Olds T, Kantama L, de Jong H. Cytogenet Genome Res. 2004;106:173–183. doi: 10.1159/000079284. [DOI] [PubMed] [Google Scholar]
- 11.Matzk F, Meister A, Schubert I. Plant J. 2000;21:97–108. doi: 10.1046/j.1365-313x.2000.00647.x. [DOI] [PubMed] [Google Scholar]
- 12.Voigt M-L, Melzer M, Rutten T, Mitchell-Olds T, Sharbel TF. In: Apomixis: Evolution, Mechanisms and Perspectives. Hörandl E, Grossniklaus U, Van Dijk P, Sharbel TF, editors. Germany: Koeltz, Koenigstein; 2007. pp. 235–258. [Google Scholar]
- 13.Schranz ME, Dobeš C, Koch MA, Mitchell-Olds T. Am J Bot. 2005;92:1797–1810. doi: 10.3732/ajb.92.11.1797. [DOI] [PubMed] [Google Scholar]
- 14.De Wet J. Evolution (Lawrence, Kans) 1968;22:394–397. doi: 10.1111/j.1558-5646.1968.tb05906.x. [DOI] [PubMed] [Google Scholar]
- 15.Comai L, Madlung A, Josefsson C, Tyagi A. Philos Trans R Soc London B. 2003;258:1149–1155. doi: 10.1098/rstb.2003.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madlung A, Comai L. Ann Bot (London) 2004;94:481–495. doi: 10.1093/aob/mch172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge RW, Martienssen R, Comai L. Plant J. 2005;41:221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- 18.Mittelstein Scheid O, Afsar K, Paszkowski J. Nat Genet. 2003;34:450–454. doi: 10.1038/ng1210. [DOI] [PubMed] [Google Scholar]
- 19.Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee H-S, Comai L, Madlung A, Doerge RW, Colot V, Martienssen RA. Trends Genet. 2003;18:141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 20.Riddle NC, Birchler JA. Trends Genet. 2003;19:597–600. doi: 10.1016/j.tig.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. Proc Natl Acad Sci USA. 2006;103:6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schranz ME, Kantama L, de Jong H, Mitchell-Olds T. New Phytol. 2006;171:425–438. doi: 10.1111/j.1469-8137.2006.01765.x. [DOI] [PubMed] [Google Scholar]
- 23.Akiyama Y, Hanna WW, Ozias-Akins P. Theor Appl Genet. 2005;111:1042–1051. doi: 10.1007/s00122-005-0020-5. [DOI] [PubMed] [Google Scholar]
- 24.Akiyama Y, Conner JA, Goel S, Morishige DT, Mullet JE, Hanna WW, Ozias-Akins P. Plant Physiol. 2004;134:1733–1741. doi: 10.1104/pp.103.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goel S, Chen Z, Conner JA, Akiyama Y, Hanna WW, Ozias-Askins P. Genetics. 2003;163:1069–1082. doi: 10.1093/genetics/163.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schranz ME, Windsor AJ, Song B-H, Lawton-Rauh A, Mitchell-Olds T. Plant Physiol. 2007;144:286–298. doi: 10.1104/pp.107.096685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jablonka E. BioEssays. 2004;26:1327–1332. doi: 10.1002/bies.20140. [DOI] [PubMed] [Google Scholar]
- 28.Camacho JPM, Sharbel TF, Beukeboom LW. Philos Trans R Soc London B. 2000;355:163–178. doi: 10.1098/rstb.2000.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachtrog D, Charlesworth B. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- 30.Roche D, Hanna WW, Ozias-Akins P. Sexual Plant Reprod. 2001;13:343–349. [Google Scholar]
- 31.Green DM. Genome. 1990;33:818–824. [Google Scholar]
- 32.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grewal SI, Moazed D. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 34.Zhong XB, De Jong JH, Zabel P. Chromosome Res. 1996;4:24–28. doi: 10.1007/BF02254940. [DOI] [PubMed] [Google Scholar]