Abstract
Mutants that retain greenness of leaves during senescence are known as “stay-green” mutants. The most famous stay-green mutant is Mendel's green cotyledon pea, one of the mutants used in determining the law of genetics. Pea plants homozygous for this recessive mutation (known as i at present) retain greenness of the cotyledon during seed maturation and of leaves during senescence. We found tight linkage between the I locus and stay-green gene originally found in rice, SGR. Molecular analysis of three i alleles including one with no SGR expression confirmed that the I gene encodes SGR in pea. Functional analysis of sgr mutants in pea and rice further revealed that leaf functionality is lowered despite a high chlorophyll a (Chl a) and chlorophyll b (Chl b) content in the late stage of senescence, suggesting that SGR is primarily involved in Chl degradation. Consistent with this observation, a wide range of Chl–protein complexes, but not the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit, were shown to be more stable in sgr than wild-type plants. The expression of OsCHL and NYC1, which encode the first enzymes in the degrading pathways of Chl a and Chl b, respectively, was not affected by sgr in rice. The results suggest that SGR might be involved in activation of the Chl-degrading pathway during leaf senescence through translational or posttranslational regulation of Chl-degrading enzymes.
Keywords: locus, pea, rice, senescence, stay green
Leaf yellowing is one of the most prominent characteristics of plant senescence, which is caused by unmasking of preexisting carotenoid by chlorophyll (Chl) breakdown (1). Chl plays a central role in light energy absorption during photosynthesis, existing mostly as Chl–protein complexes. Higher plants contain two species of Chls, Chl a and Chl b. Chl a is the major Chl and is contained in various Chl–protein complexes including photosystem I (PSI) and photosystem II (PSII) reaction center complexes and the cytochrome b6f complex. Light-harvesting complexes I and II (LHCI and LHCII, respectively) bind both Chl a and Chl b and are the only Chl b-containing protein complexes. Chl degradation plays an important role in the degradation of Chl–protein complexes, at least in LHCII (2, 3). The first step of Chl a degradation is dephytylation by chlorophyllase (Chlase) followed by Mg2+ removal from the resultant chlorophyllide a (Chide a), resulting in formation of pheophorbide a (Pheide a), the last green compound in the Chl-degrading pathway. Pheide a is then converted into red chlorophyll catabolite (RCC) by Pheide a oxygenase (PaO). In Chl b degradation, Chl b is converted into Chl a and metabolized in the Chl a-degrading pathway described above. The first step of this conversion is catalyzed by Chl b reductase.
Mutants that retain greenness of leaves during senescence are known as “stay-green” mutants, of which there are several types (4). Several genes regulating the process of leaf senescence have been identified, including genes involved in hormonal and proteosomal regulation (5, 6). Nonfunctional-type stay-green mutants primarily affecting Chl degradation, lls1/acd1, nyc1, and sgr, have also been reported. lls1/acd1 was isolated as a lesion mimic mutant but shows a stay-green phenotype during senescence under dark conditions (7–9). LLS1/ACD1 encodes PaO, and Pheide a accumulates in senescent leaves of lls1/acd1. Free Chls and their colored catabolites generate reactive oxygen species (ROS) under light conditions, suggesting that the generation of ROS with the accumulation of Pheide a in lls1/acd1 leads to cell death, resulting in the lesion mimic phenotype (2). NYC1 in rice encodes a probable Chl b reductase (3), whereas defects in NYC1 result in stabilization of the LHCII and photosynthetic pigments in LHCII, including Chl a and carotenoids as well as Chl b. On the other hand, SGR is thought to be localized in plastids but does not show significant similarity to any other protein with a known function (2, 10, 11).
Gregor Mendel determined the law of genetics by using seven pairs of traits in garden pea: cotyledon color, seed shape, pod color, pod shape, flower color, flower position, and stem length (12). Seed shape (smooth versus wrinkled) and stem length are regulated by the R and Le loci and correspond to natural mutations of a starch-branching enzyme gene (13) and a gibberellin 3β-hydroxylase gene (14, 15), respectively. Cotyledon color (yellow versus green) is regulated by the I locus (16), with the i mutant retaining not only greenness of the cotyledon during seed maturation but also greenness of leaves during senescence, suggesting that this mutant is a stay-green mutant (17). Recently, Armstead et al. (11) showed linkage between the I locus and pea homolog of SGR. Here, we present convincing evidence that Mendel's green cotyledon gene is the SGR in pea (PsSGR). Furthermore, we report the functional analysis of the SGR in rice (OsSGR). OsSGR was shown to be strongly expressed in senescent leaves and involved in activation of the Chl-degrading pathway through translational or posttranslational regulation of Chl-degrading enzymes.
Results and Discussion
Mendel's Green Cotyledon Mutation Is a Stay-Green Mutation.
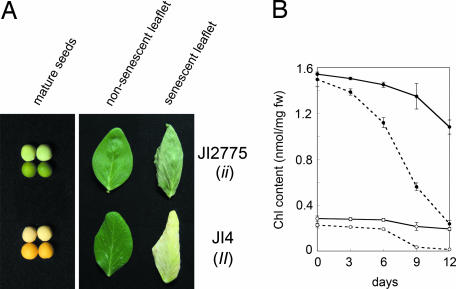
The pea strain JI2775 has a defect on the I locus and presents green cotyledons in mature seeds (Fig. 1A). JI2775 also retains greenness of leaves during leaf senescence, suggesting that JI2775 is a stay-green mutant and that the I gene functions in Chl degradation both in cotyledons during seed maturation and in true leaves during senescence (Fig. 1A). Twenty-five green cotyledon plants from the segregating F2 population between JI2775 and wild type (JI4) did not show yellowing during senescence, confirming that these two traits are caused by the mutation in the I locus. We measured the Chl contents of JI4 and JI2775 during dark-induced leaf senescence (Fig. 1B), revealing that both Chl a and Chl b contents in JI4 drastically decreased by 12 days of dark incubation (DDI). In contrast, JI2775 retained 70% and 68% of the initial content of Chl a and b at 12 DDI, respectively; that is, the contents of both Chl a and Chl b were retained at a higher level in JI2775, suggesting that the I gene is involved in the degradation mechanisms of both Chl a and Chl b.
Fig. 1.
Stay-green phenotype of the i mutant in pea. (A) Seed color phenotype and naturally senescent leaves of JI4 (II) and JI2775 (ii). (Left) mature seeds with (Upper) and without (Lower) seed coats. (Right) Nonsenescent (Left) and naturally senescent (Right) leaves. Some parts of the senescent leaflet of JI4 and JI2775 are dead, suggesting that both leaflets are in the very late stage of leaf senescence. JI2775 retained greenness even at this stage. (B) Change in Chl content in pea during dark incubation. Detached leaves of the same fresh weight were extracted with the same volume of 80% acetone, and Chl contents were measured spectrophotometrically. Solid line, JI2775; dotted line, JI4; filled circles, Chl a; open circles, Chl b. Bars indicate standard errors; n = 3.
The Fv/Fm (the ratio of variable to maximum chlorophyll florescence) value represents PSII activity and is one of the parameters of leaf functionality. Chl contents of nonsenescent leaves with high Fv/Fm values (for example, higher than 0.81) were similar between JI4 and JI2775 [supporting information (SI) Fig. 7]. On the other hand, naturally senescent leaves of JI2775 with an Fv/Fm value lower than 0.78 retained a much higher Chl content than JI4 (SI Fig. 7). This observation suggests that JI2775 is a nonfunctional stay-green mutant.
Tight Linkage Between PsSGR and the I Locus.
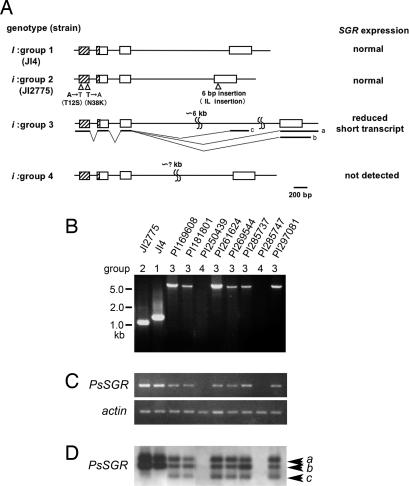
To date, three nonfunctional stay-green genes, PaO, NYC1, and SGR, have been reported (2, 3, 7–10). We raised an F2 population between JI4 and JI2775 and analyzed the linkage between the pea homologs of these stay-green genes and the cotyledon color phenotype in mature seeds. A partial genomic sequence of PaO in pea was obtained, revealing a polymorphism between JI4 and JI2775, but no linkage was found with the cotyledon color phenotype (data not shown). Next, we isolated NYC1 in pea but found no sequence difference between JI4 and JI2775, at least not in the exons and introns (data not shown). Then we examined the SGR in pea (PsSGR). Degenerate PCR was performed to clone a partial sequence of PsSGR, and a full-length sequence was isolated by using inverse PCR and 3′-RACE. Analysis of the PsSGR genome sequence revealed that PsSGR consists of four exons and three introns and has a sequence length polymorphism of the third intron between JI4 and JI2775 (Fig. 2A). We analyzed 93 green-cotyledon (ii) and 195 yellow-cotyledon (II or Ii) seeds from the F2-segregating population between JI4 and JI2775 and found no recombinants between the cotyledon color phenotype and intron length polymorphism, confirming that the I locus and PsSGR are linked very tightly (11).
Fig. 2.
Molecular analysis of the i alleles. (A) Genomic structures of four groups of PsSGR. Group 1 is the wild type (I) and groups 2–4 are mutants (i). Boxes represent exons, and lines are UTRs and introns. Hatched regions correspond to the transit peptide. Bars a, b, and c under the group 3 genome structure indicate the three kinds of transcripts observed in group 3. In group 2, two amino acid substitutions and a two-amino acid insertion were found. The expression level and splicing were normal. In group 3, the third intron was very long, and a short transcript, shown as transcript c, derived from a cryptic splice site, was detected. Transcripts a and b differ in the length of 3′-UTR, and these transcripts were also observed in other groups (see D). The expression level was reduced (see C). In group 4, no amplification of the third intron was observed in genomic PCR, although all exons were retained. mRNA was not detected by RT-PCR analysis (see C). (B) Genomic PCR analysis of the third intron. The strain name and corresponding group are shown above each lane. (C) Semiquantitative RT-PCR analysis of PsSGR. RNA was extracted from leaves incubated in the dark at 18°C for 10 days. The amplified PCR product corresponded to the second to forth exons. Actin was amplified as a reference. (D) RT-PCR Southern blot analysis of the 3′-RACE products using the same RNA as in C. Arrowheads a, b, and c correspond to transcripts a, b, and c in A.
The I Gene Is the SGR in Pea.
The SGR does not show significant similarity to any protein whose function is known, but it is highly conserved within the plant kingdom (ref. 11 and SI Fig. 8). Expression of PsSGR was induced in dark-induced senescence (SI Fig. 9A). Further, splicing of PsSGR occurred identically, and the expression levels were similar between JI4 and JI2775 during dark-induced and natural senescence (Fig. 2C and SI Fig. 9). Armstead et al. (11) reported that the RNA level of PsSGR is very low in JI2775, but this observation was in contrast to ours. We found several amino acid differences between PsSGRJI2775 and PsSGRJI4: Thr to Ser at amino acid 12 (T12S), Asp to Lys at amino acid 38 (N38K), and a two-amino acid insertion of Ile and Leu at 189 (Fig. 2A and SI Fig. 8), suggesting that if a mutation in PsSGR is responsible, one of these amino acid differences is responsible for the stay-green phenotype in JI2775. T12S and N38K substitutions are located in the transit peptide according to TargetP (www.cbs.dtu.dk/services/TargetP) (SI Fig. 8). To examine whether these amino acid substitutions affect the intracellular localization of PsSGR, we introduced constructs with fusion of the green fluorescent protein (GFP) to PsSGRJI4 or PsSGRJI2775 into onion epidermal cells by particle bombardment (SI Fig. 10). The signals for both constructs were colocalized with that of the red fluorescent protein (RFP) fused to the transit peptide from the plastid-localizing rice S9 ribosomal protein (18), suggesting that both PsSGRJI4 and PsSGRJI2775 are localized in plastids, and the two amino acid substitutions in the PsSGRJI2775 transit peptide are not responsible for the stay-green phenotype of JI2775. The two-amino acid insertion is located in a relatively conserved region of the SGR proteins, and there is no length difference in this region even between dicotyledous and monocotyledous SGR proteins (SI Fig. 8). To examine whether the amino acid insertions impair SGR function, we introduced a PsSGRJI4 coding sequence (CDS) driven by the rice SGR (OsSGR) promoter into a rice sgr mutant (see below); however, it did not complement the sgr phenotype well, probably because of the distant relationship between PsSGR and OsSGR (they show 60.7% amino acid identity). We therefore made a construct harboring OsSGR cDNA with a 6-bp insertion (corresponding to Ile and Leu) at the site corresponding to PsSGRJI2775 (at amino acid 194 in OsSGR) and introduced it into a rice sgr mutant under control of the OsSGR promoter. Wild-type OsSGR restored the sgr stay-green phenotype, but the mutated OsSGR did not (SI Fig. 11), suggesting that the two-amino acid insertion is responsible for the defect in SGR function in JI2775.
Genetic complementation tests with JI2775 newly identified eight i mutant lines (Fig. 2A), and PCR analysis corresponding to the third intron revealed four types of polymorphism (Fig. 2B): group 1 with a 1.4-kb PCR product corresponding to JI4 (wild type); group 2 with a 1.2-kb PCR product corresponding to JI2775; group 3 with a very large, ≈6.5-kbp PCR product; and group 4 with no amplification. Full-length SGR mRNA of group 3 had the same sequence as that of group 1 (JI4). In addition, semiquantitative RT-PCR analyses revealed that group 3 showed reduced expression and group 4 showed no expression of PsSGR (Fig. 2C). Southern blot analysis of the 3′-RACE products revealed a short mRNA product in group 3 (Fig. 2D). The short transcript was generated by incorrect splicing of the third intron, resulting in a truncated 160-amino acid protein with an additional 3 amino acids of Val-Gln-Asp (Fig. 2A and SI Fig. 8). It is possible that this truncated SGR product might have a weak dominant-negative effect on the normal SGR. Based on genomic PCR analysis, group 4 PsSGR were shown to have all exons (data not shown), suggesting that the lack of amplification of the region containing the third intron might be because the third intron of group 4 was too long for PCR amplification or contains a sequence that could not be amplified easily. The lack of PsSGR expression in group 4 suggests that group 4 PsSGR has a transcriptional defect, which causes the stay-green phenotype. All i locus mutant lines analyzed had defects in PsSGR, confirming that the I locus encodes PsSGR.
Functional Characterization of SGR in Rice.
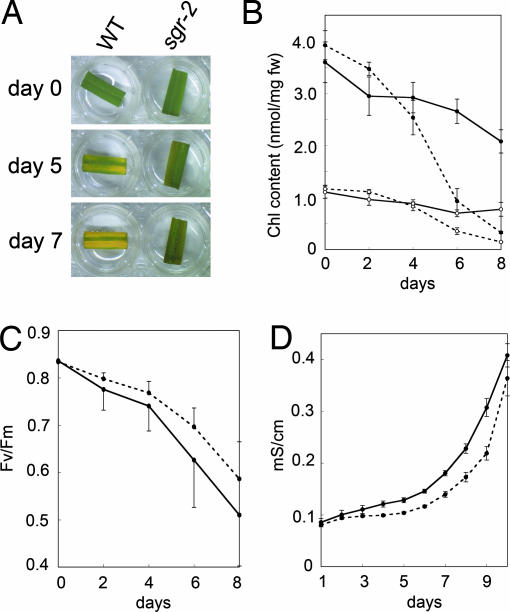
For further investigation of the function of SGR, we examined rice because analysis of gene function is easier, and more information is available for rice than pea. In a screen of stay-green mutants in rice, we found a mutant harboring an 8-bp deletion 55 bp from the initiation codon in the first exon, and it was designated sgr-2. This deletion caused a frameshift, which generated a predicted protein lacking most of the conserved amino acid sequences in SGR, suggesting that sgr-2 is a null mutant. In fact, sgr-2 showed a stay-green phenotype during dark-induced senescence (Fig. 3A). As described above, sgr-2 plants containing a full-length SGR CDS driven by the OsSGR promoter showed yellowing during dark-induced senescence, confirming that this gene is responsible for the stay-green phenotype in sgr-2 (SI Fig. 11). sgr-2 retained 58% and 66% of the initial Chl a and b contents, respectively, at the very late stage of senescence (8 DDI), indicating that sgr affects the degradation of both Chl a and Chl b (Fig. 3B). The Fv/Fm value decreased in sgr-2 in a manner similar to the wild type despite the high Chl content in sgr-2 during dark-induced senescence (Fig. 3C). These observations are basically identical to those observed with the i mutation in pea. In addition, the increase in membrane ion leakage was not delayed in sgr-2 (Fig. 3D), confirming that sgr is a nonfunctional stay-green mutant.
Fig. 3.
Physiological analysis of senescence in the rice sgr mutant. (A) Dark-induced leaf senescence in sgr-2. (B) Change in Chl content during dark incubation in rice. Chl contents were measured as in Fig. 1B. Solid line, sgr-2; dotted line, WT; filled circles, Chl a; open circles, Chl b. Bars indicate standard errors; n = 3. (C) Fv/Fm values. Solid line, sgr-2; Dotted line, WT. Bars indicate standard errors; n = 3. (D) Membrane ion leakage. Solid line, sgr-2; dotted line, WT. Bars indicate standard errors; n = 11.
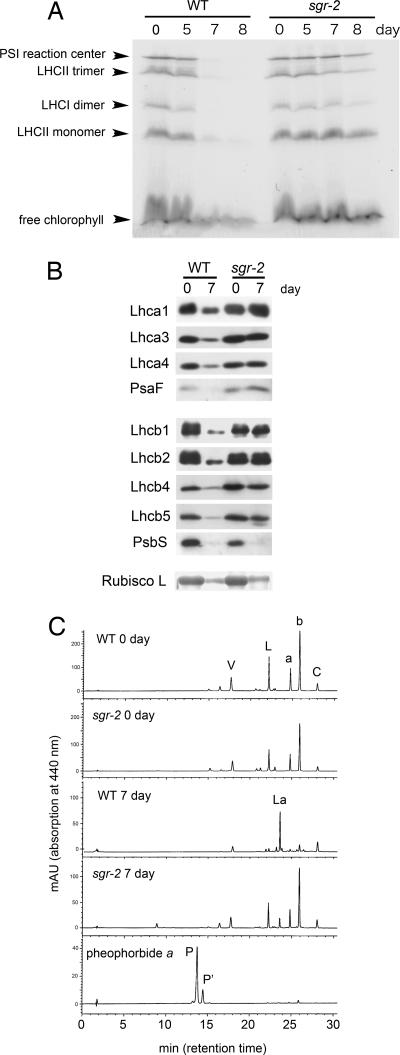
sgr-2 showed a distinct phenotype compared with nyc1, another nonfunctional stay-green mutant in rice. In nyc1, degradation of both Chl a and Chl b is inhibited, but retention of Chl b is predominant, and the Chl a/b ratio is close to 1 at the very late stage of senescence (3). This Chl a/b ratio is the result of the selective retention of LHCII at the late stage of senescence in nyc1, because LHCII contains both Chl a and Chl b at a ratio of 1.3. sgr-2 retained a higher Chl content and showed a higher Chl a/b ratio (2.68 at 8 DDI) than nyc1 at the same stage. To examine the retention of Chl–protein complexes in sgr-2 during senescence, “green-gel” analysis was performed (Fig. 4A). LHCII trimer and monomer bands were retained in sgr-2 as well as nyc1 during senescence. In addition, bands for the PSI reaction center complex, which contains only Chl a, and LHCI dimer, both of which disappear during senescence in nyc1 (3), were more stable in sgr-2 than the wild type. The observation that Chl a–protein complexes as well as LHCI and LHCII are stable is consistent with the fact that both Chl a and Chl b are more stable in sgr than the wild type during senescence. Western blot analysis revealed that the LHCI and LHCII isoforms analyzed were more stable in sgr-2 than the wild type during senescence (Fig. 4B). However, not all Chl–protein complexes appeared stable in sgr-2, such as PsbS. SDS/PAGE analysis showed that a stroma protein, the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit, is degraded normally in sgr-2, suggesting that SGR is involved in the degradation of a wide range of Chl–protein complexes but not in the degradation of stroma proteins, or at least not the Rubisco large subunit.
Fig. 4.
Protein and pigment degradation during senescence in the rice sgr mutant. (A) Change in Chl-binding proteins during dark incubation. The gel was visualized by Chl bound to proteins (green gel). (B) Western blot analysis of photosynthetic proteins from nonsenescent (0 DDI) and senescent (7 DDI) leaves. Rubisco large subunit (Bottom) was detected by SDS/PAGE analysis with Coomassie brilliant blue G 250. (C) HPLC elution profiles of photosynthetic pigments extracted from the rice leaves measured at 440 nm. Extracts from nonsenescent (0 day) and fully senescent (7 day) leaves of WT and sgr-2 were analyzed. Pheophorbide a indicates the HPLC elution profile of commercially available pheophorbide a. V, violaxanthin; L, lutein; La, lutein 3-acetate; b, Chl b; a, Chl a; C, β-carotene; P, Pheide a; P′, an epimer of Pheide a; AU, absorption unit.
Photosynthetic pigment composition in sgr-2 was analyzed by HPLC (Fig. 4C). The composition of photosynthetic pigments in sgr-2 was basically identical to that of the wild type before senescence (0 DDI). At the late stage of senescence (7 DDI), sgr-2 retained more Chl a and Chl b than the wild type as shown in Fig. 3B. Although most lutein was converted into lutein 3-acetate at the late stage of senescence in the wild type (M.K., T. Maoka, and S. Takaichi, unpublished observations), full conversion did not occur in sgr-2 and nyc1 (Fig. 4C and ref. 3). Lutein in LHCI and LHCII may not be converted into lutein 3-acetate in sgr, as lutein in LHCII is not converted in nyc1 (3). No significant accumulation of Pheide a was detected in sgr-2 (Fig. 4C and SI Fig. 12), suggesting that the greenness of senescent sgr leaves is not directly related to the accumulation of Pheide a. Accumulation of Pheide a results in a lesion mimic phenotype as demonstrated in the PaO mutants lls1 and acd1 (7, 8, 19). sgr-2 did not show a lesion mimic phenotype in light conditions (data not shown), consistent with the lack of significant accumulation of Pheide a in sgr-2. In addition, Pheide a did not accumulate in the cotyledon of JI2775 mature seeds (data not shown). These observations suggest that Chl retention in sgr is caused by inhibition of the first step of Chl a and Chl b degradation rather than feedback inhibition of Chl a and Chl b degradation with the accumulation of Pheide a.
SGR Is Not Involved in Expression of Chl-Degrading Enzyme Genes.
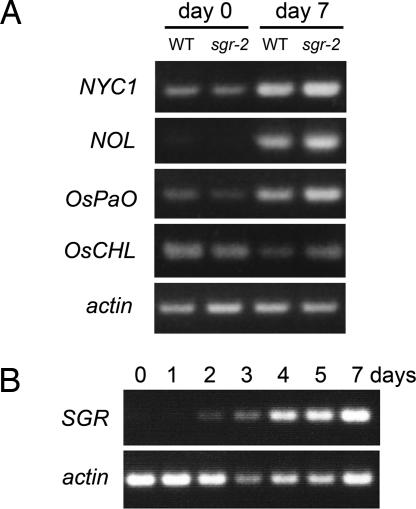
Whether SGR is involved primarily in the stability of Chls or Chl-containing proteins is controversial. Prior degradation of Chl b is required for LHCII degradation (3), suggesting that in the degradation process of LHCII, SGR primarily functions in Chl b degradation. SGR is thought to directly or indirectly regulate Chl b degradation through the probable Chl b reductase NYC1 because NYC1 is thought to be an enzyme that metabolizes Chl b directly, and a null mutation in NYC1 resulted in a severe Chl b degradation defect. In sgr-2, transcription of NYC1 was induced at a level comparable with that of the wild type in senescent leaves (Fig. 5A), suggesting that SGR or an SGR-mediated pathway regulates the function of NYC1 in a translational or posttranslational manner (Fig. 5A). Such regulation might include regulation of protein synthesis/stability, enzyme activity, and accessibility to substrate pigments. Another Chl b reductase gene, NOL, which is thought to play a minor role in Chl b degradation during leaf senescence (3), was also expressed in sgr-2 at a level comparable with that of the wild type, suggesting that SGR is not involved in NOL expression (Fig. 5A).
Fig. 5.
Expression of SGR and the genes encoding enzymes in the Chl-degrading pathway. (A) Semiquantitative RT-PCR analysis of the genes encoding enzymes in the Chl-degrading pathway in dark-induced senescent leaves. (Bottom) Expression of actin as a reference. (B) Semiquantitative RT-PCR analysis of SGR expression during dark-induced senescence. (Bottom) Expression of actin as a reference.
The role of SGR in Chl a degradation was less evident. Because Chl a degradation was obviously inhibited in sgr-2, it is possible that SGR regulates the function of Chlase, the first enzyme in the Chl a-degrading pathway. Transcript level of Chlase in rice (OsCHL) in sgr-2 was comparable with that in the wild type, suggesting that SGR is not involved in transcriptional regulation of these genes (Fig. 5A). Recently, it was reported that the in vivo function of Chlase is thought to be regulated in a posttranslational manner (20). SGR or an SGR-mediated pathway might therefore regulate the activity of Chlase posttranslationally. Alternatively, the degradation of Chl a–protein complexes by proteases might be required to degrade Chl a. In this scenario, SGR or an SGR-mediated pathway may regulate Chl a protein–complex-degrading proteases.
Furthermore, expression level of PaO in rice (OsPaO) in sgr-2 was also comparable with that in the wild type (Fig. 5A). Very recently, Ren et al. (21) reported that a mutant in NYE1, an SGR orthlog in Arabidopsis thaliana, have reduced PaO activity. SGR might activate the Chl-degrading pathway during senescence via translational/posttranslational regulation of enzymes in the Chl-degrading pathway including Chl b reductase, Chlase, and PaO.
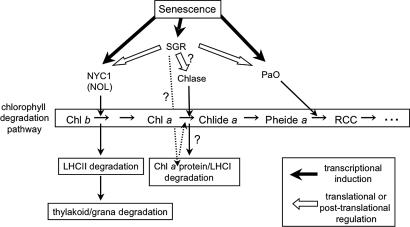
In summary, linkage analysis and the molecular characterization of PsSGR in three independent i alleles confirmed that the I gene is PsSGR. SGR is thought to be involved in Chl degradation through translational/posttranslational regulation of Chl-degrading enzymes. Senescence signals activate the Chl-degrading pathway, at least partly via SGR, whose expression is also induced by senescence signals (Fig. 5B). The degradation of Chl induces downstream events such as LHCII degradation and thylakoid/grana degradation (Fig. 6). Consistent with this model, sid, an sgr mutant in Festuca pratensis, shows more stable LHCII and grana structures than the wild type during senescence (11, 22, 23).
Fig. 6.
Model of SGR function during leaf senescence. NYC1, NOL, Chlase, and PaO are the enzymes in the Chl-degrading pathway. Chl b is converted into Chl a and metabolized in the Chl a-degrading pathway. The first step of this conversion is catalyzed by the Chl b reductase NYC1. Chl b degradation is necessary for LHCII degradation and thylakoid/grana degradation in senescent leaves. NOL, another Chl b reductase, may also have a minor role in Chl b degradation during leaf senescence (3). Chlase forms Chlide a by dephytylation of Chl a, the major Chl species in higher plants. PaO converts Pheid a into the nongreen compound RCC. Senescence signals induce expression of NYC1, NOL, SGR, and PaO (bold arrows). SGR is involved in Chl b degradation and the resultant LHCII degradation via translational/posttranslational regulation of NYC1 (white arrows). Similarly, SGR might be involved in Chl a degradation via posttranslational regulation of Chlase (white arrows) because posttranslational regulation may be important for in vivo activity of Chlase (20). Alternatively, SGR might be involved in Chl a degradation via the regulation of Chl a-containing protein stability (dashed arrows). In addition, PaO activity is regulated by SGR at the translational/posttranslational level (21).
Materials and Methods
Plant Materials and Growth Conditions.
Pea strains, JI4 and JI2775, were obtained from the John Innes Centre Public Collection (Norwich, U.K.), and other green cotyledon pea lines (PI169608, PI181801, PI250439, PI261624, PI285717, PI285737, PI285747, PI297081) were from the U.S. Department of Agriculture (Washington, D. C.). sgr-2 was isolated from our collection of stay-green mutant lines by using TILLING-based screening (24, 25). sgr-2 was derived from the rice cultivar Nipponbare irradiated with carbon ions (220 MeV) at Takasaki Advanced Radiation Research Institute, Japan Atomic Energy Agency (Tokyo, Japan) (26). Nipponbare was used as the wild-type rice in this work. Dark incubation was performed at 18°C in pea and 27°C in rice.
Analysis of Photosynthetic Pigments, Photochemical Efficiency, and Membrane Ion Leakage.
For pigment extraction, plant tissues of the same fresh weight were ground in a mortar with liquid nitrogen and extracted with the same volume of 80% acetone. The HPLC apparatus used for analysis was equipped with a Symmetry C8 column (Waters, Milford, MA) and a photodiode-array detector (SPD-M10A; Shimadzu, Kyoto, Japan) according to Kusaba et al. (3). Pehide a was purchased from Wako Pure Chemical Industries (Osaka, Japan). Chl a and Chl b contents were determined according to Porra et al. (27). Fv/Fm values were measured with an OS1-FL fluorometer (Opti-Sciences, Hudson, NH) according to the manufacturer's instructions. For measurement of membrane ion leakage, three leaf discs of 6-mm diameter floated on 500 μl of water were incubated in the dark. Conductivity was measured with a Twin Cond B-173 conductivity meter (Horiba, Kyoto, Japan).
Isolation of the PsSGR Gene.
Degenerate PCR was performed by using primers gtiytittyytiggigtigayga and cciswdatrtgrcartgiacrtg, corresponding to conserved amino acid sequences between AtSGR and OsSGR, with cDNA prepared from senescent leaves as a template. 3′-RACE was performed by using first-strand cDNA primed with oligo(dT) with an adaptor (gtcatctagaggtaggccatggactgaggtaaccttttttttttttttttt), and the product was used as a template for PCR with primer pairs PsSGR-1F (aaacaaacacccaggaaatcttccg), derived from the degenerate PCR product sequence, and adaptor 1 (gtcatctagaggtaggccatgg). Pea genomic DNA was digested with EcoRV then ligated and used for inverse PCR (first PCR primers, agttgtttatggtttgagaaattgcc and gtgcacgatcatatctcagtattg; second PCR primers, ggtaacatcactatgtgttaac and tgctacaaacatgtgaattgattgc). To obtain the full-length CDS of PsSGR, PsSGR-5′-UTR-F (gaataataagcacgtaaccttggtg) and PsSGR-3′-UTR-R (attccaattttaaaccatacaaatgtc) were used.
Protein and RNA Analyses.
Green-gel analysis, SDS/PAGE, and Western blot analysis were performed according to Morita et al. (28). For green-gel analysis, 100-mg fresh weight leaf samples were extracted with 250 μl of extraction buffer [0.3 M Tris, pH 6.8/1% SDS/2% Triton X-100/10% (vol/vol) glycerol]. For SDS/PAGE and Western blot analysis, 100-mg fresh weight leaf samples were extracted with 2 ml of 1× SDS buffer [62.5 mM Tris, pH 6.8/2.5% SDS/5% mercaptoethanol/10% (vol/vol) glycerol]. Antibodies against Lhcas, Lhcbs, and PsbS were purchased from Agrisera (Vännäs, Sweden), and anti-PsaF antibody was provided by Y. Takahashi (Okayama University, Okayama, Japan). In the semiquantitative RT-PCR, first strand cDNAs were synthesized from 3 μg of total RNA. Primers used for amplification of pea and rice genes are listed in SI Table 1. In DNA gel blot analysis of the 3′-RACE products and quantitative RT-PCR, 5 μl of PCR products was electrophoresed in 1.0% agarose gel, transferred to a nylon membrane, and detected with a digoxigenin labeling system (Roche Diagnostics, New York, NY) by using the full-length PsSGR CDS as a probe. For real-time PCR analysis and RT-PCR DNA gel blot analysis, cDNA was synthesized by using 2 μg of total RNA from pea leaves at 10 DDI and random hexamer primers. For PsSGR, PCR primers tgggacccactctgatttg and cggtggaaaacaacactcac and Universal ProbeLibrary 43 (Roche Diagnostics) were used, and for actin in pea, PCR primers gtctgtgacaatggaactggaatg and gtctcaaacatgatttgggtcatc and Library 9 were used. Amplification and expression quantification were performed by using the ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA) and FastStart TaqMan probe master (Roche Diagnostics).
Construction and Visualization of the GFP Fusion Protein.
A plastid-localized RFP construct with the transit peptide from rice ribosomal protein S9 is the RFP derivative of OsPRS9TP-GFP (18). PsSGRJI2775-GFP and PsSGRJI4-GFP were constructed by insertion of the full-length PsSGR CDS fragment into the XbaI–BamHI site of pJ4-GFP (29). Particle bombardment was performed with a PDS-1000/He particle gun (Bio-Rad, Hercules, CA). Three micrograms of plasmid precipitated onto 1.0-μm gold beads was introduced into onion epidermal cells, then 24 h after bombardment, images of GFP and RFP signals were captured with a BZ-8000 fluorescence microscope (Keyence, Osaka, Japan). The filters used were OP-66836 for GFP and OP-66838 for RFP.
Amino Acid Alignment.
Amino acid sequences of the SGR were aligned by using ClustalW (www.ddbj.nig.ac.jp/search/clustalw-j.html) and were then adjusted manually.
Rice Transformation.
The promoter region of OsSGR was isolated by PCR with primers ggtaccggacggaattcacctctag and gggtctgctccctcggatctcttagtc and used for plasmid construction. A binary vector with the OsSGR promoter, CDS of OsSGR, and Nos terminator was transformed into Agrobacterium tumefaciens strain EHA101. Rice transformation was conducted according to Yokoi et al. (30).
Supplementary Material
Acknowledgments
We thank Yoshihiro Hase, Naoya Shikazono, and Atsushi Tanaka for irradiating rice seeds with carbon ions; Mike Ambrose (John Innes Centre) for providing pea strains JI4 and JI2775; Yuichiro Takahashi (Okayama University) for providing the anti-PsaF antibody; Toshio Takyu for helpful suggestions, and Yasuo Nagato for encouragement. This work was supported by a grant for nuclear research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, a grant from the Research Fellowships for Young Scientists, and a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Rice Genome Project IP-1011).
Abbreviations
- CDS
coding sequence
- Chide
chlorophyllide
- Chl
chlorophyll
- Chlase
chlorophyllase
- DDI
days of dark incubation
- Fv/Fm
ratio of variable to maximum chlorophyll florescence
- LHCI
light-harvesting complex I
- LHCII
light-harvesting complex II
- PaO
Pheide a oxygenase
- Pehide
pheophorbide
- PSI
photosystem I
- PSII
photosystem II
- RCC
red chlorophyll catabolite
- RFP
red fluorescent protein
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AB303331 (PsSGR; JI4), AB303332 (PsSGR; JI2775), AAW82959 (GmSGR1), AAW82960 (GmSGR2), AAY98500 (LeSGR), AY850161 (AtSGR1), AY699948 (AtSGR2), AAW82958 (SbSGR), AAW82956 (ZmSGR1), AAW82957 (ZmSGR2), AAW82954 (OsSGR), AAW82955 (HvSGR), BAF49740 (NYC1), BAF49741 (NOL), AP008216 (OsCHL), and AK120554 (OsPaO)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0705521104/DC1.
References
- 1.Matile P. Exp Gerontol. 2000;35:145–158. doi: 10.1016/s0531-5565(00)00081-4. [DOI] [PubMed] [Google Scholar]
- 2.Hörtensteiner S. Annu Rev Plant Biol. 2006;57:55–77. doi: 10.1146/annurev.arplant.57.032905.105212. [DOI] [PubMed] [Google Scholar]
- 3.Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. Plant Cell. 2007;19:1362–1375. doi: 10.1105/tpc.106.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas H, Howarth J. J Exp Bot. 2000;51:329–337. doi: 10.1093/jexbot/51.suppl_1.329. [DOI] [PubMed] [Google Scholar]
- 5.Lim PO, Woo HR, Nam HG. Trends Plant Sci. 2003;8:272–278. doi: 10.1016/S1360-1385(03)00103-1. [DOI] [PubMed] [Google Scholar]
- 6.Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. Proc Natl Acad Sci USA. 2006;103:814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray J, Close PS, Briggs SP, Johal GS. Cell. 1997;89:25–31. doi: 10.1016/s0092-8674(00)80179-8. [DOI] [PubMed] [Google Scholar]
- 8.Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S. Proc Natl Acad Sci USA. 2003;100:15259–15264. doi: 10.1073/pnas.2036571100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka R, Hirashima M, Satoh S, Tanaka A. Plant Cell Physiol. 2003;44:1266–1274. doi: 10.1093/pcp/pcg172. [DOI] [PubMed] [Google Scholar]
- 10.Cha KW, Lee YJ, Koh HJ, Lee BM, Nam YW, Paek NC. Theor Appl Genet. 2002;104:526–532. doi: 10.1007/s001220100750. [DOI] [PubMed] [Google Scholar]
- 11.Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. Science. 2007;315:73. doi: 10.1126/science.1132912. [DOI] [PubMed] [Google Scholar]
- 12.Mendel G. Verh Naturforsch Ver Brünn. 1866;4:3–47. [Google Scholar]
- 13.Bhattachayya MK, Smith AM, Ellis THN, Hedey C, Martin C. Cell. 1990;60:115–122. doi: 10.1016/0092-8674(90)90721-p. [DOI] [PubMed] [Google Scholar]
- 14.Lester DR, Ross JJ, Davies PJ, Reid JB. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin DN, Proebsting WM, Hedden P. Proc Natl Acad Sci USA. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White OE. Proc Am Philos Soc. 1917;56:487–588. [Google Scholar]
- 17.Thomas H, Schellenberg M, Vicentini F, Matile P. Bot Acta. 1996;109:3–4. [Google Scholar]
- 18.Arimura S, Takusagawa S, Hatano S, Nakazono M, Hirai A, Tsutsumi N. FEBS Lett. 1999;450:231–234. doi: 10.1016/s0014-5793(99)00491-3. [DOI] [PubMed] [Google Scholar]
- 19.Yang M, Wardzala E, Johal GS, Gray J. Plant Mol Biol. 2004;54:175–191. doi: 10.1023/B:PLAN.0000028789.51807.6a. [DOI] [PubMed] [Google Scholar]
- 20.Harpaz-Saad S, Azoulay T, Arazi T, Ben-Yaakov E, Mett A, Shiboleth YM, Hörtensteiner S, Gidoni D, Gal-On A, Goldschmidt EE, et al. Plant Cell. 2007;19:1007–1022. doi: 10.1105/tpc.107.050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B. Plant Physiol. 2007;144:1429–1441. doi: 10.1104/pp.107.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas H. Planta. 1977;137:53–60. doi: 10.1007/BF00394435. [DOI] [PubMed] [Google Scholar]
- 23.Thomas H, Morgan WG, Thomas AM, Ougham HJ. Theor Appl Genet. 1999;99:92–99. [Google Scholar]
- 24.Till BJ, Burter C, Comai L, Henikoff S. Nucleic Acids Res. 2004;11:2632–2641. doi: 10.1093/nar/gkh599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato Y, Shirasawa K, Takahashi Y, Nishimura M, Nishio T. Breed Sci. 2006;56:179–183. [Google Scholar]
- 26.Naito K, Kusaba M, Shikazono N, Takano T, Tanaka A, Tanisaka T, Nishimura M. Genetics. 2005;169:881–889. doi: 10.1534/genetics.104.033654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porra RJ, Thompson WA, Kriedemann PE. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- 28.Morita R, Kusaba M, Yamaguchi H, Amano E, Miyao A, Hirochika H, Nishimura M. Breed Sci. 2005;55:361–364. [Google Scholar]
- 29.Igarashi D, Ishida S, Fukazawa J, Takahashi Y. Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoi S, Tsuchiya T, Toriyama K, Hinata K. Plant Cell Rep. 1997;16:363–366. doi: 10.1007/BF01146774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.