Abstract
The gene of Micrococcus luteus UV endonuclease (cyclobutane pyrimidine dimer–DNA glycosylase/abasic lyase) was cloned and characterized. The cloned gene, whose product had a predicted molecular mass of 17,120 Da, was found to be capable of complementing the Escherichia coli uvrA6 mutation in vivo with respect to resistance to acetone-mediated molecular photosensitization, a treatment producing exclusively cyclobutane pyrimidine dimers in DNA. It also generated a nicking activity specific for photosensitization-treated DNA by in vitro transcription/translation. When expressed in E. coli cells, the gene produced a protein structurally identical with UV endonuclease and possessing an activity consistent with cyclobutane pyrimidine dimer–DNA glycosylase/abasic lyase with respect to the effect of inhibitors and the site of the DNA backbone scission. Furthermore, the UV endonuclease-deficient mutant DB7 was shown to regain the enzyme through transformation with the cloned gene. The deduced amino acid sequence of the gene product was at best 27% identical with that of endonuclease V of phage T4, an enzyme strikingly similar to UV endonuclease in molecular and catalytic properties. Despite this marginal overall similarity in amino acid sequence, four of the seven amino acid residues reported to be functionally important in the T4 enzyme were found to be conserved in the M. luteus enzyme. We propose that the gene be called uveA.
Keywords: base excision repair, DNA damage, ultraviolet light
“UV endonuclease” of Micrococcus luteus, known as the first DNA excision repair enzyme to be isolated from cell extracts (1–3), is a cyclobutane pyrimidine dimer (CPyD)–DNA glycosylase/abasic lyase (4, 5), sharing the same combined activities with endonuclease V of phage T4. Unlike the T4 enzyme, however, UV endonuclease was slow in gaining recognition as a repair enzyme because its contribution to UV survival of the cell was obscured by a homolog of the Escherichia coli Uvr system also existing in this bacterium (6–8). Recently, it has been given a backup status in the removal of CPyDs from the cellular DNA, since a double mutant deficient in UV endonuclease and the Uvr homolog is much more sensitive to UV than a mutant deficient only in the latter (9).
Despite the early debut as an enzyme, the cloning of the UV endonuclease gene has remained elusive, making further investigation of this enzyme difficult. Meanwhile, T4 endonuclease V, which is strikingly similar to UV endonuclease not only in the substrate specificity but also in molecular properties (10), has been studied in some detail mainly from the viewpoint of structure–function relationship (11–14). Hence, information on the M. luteus counterpart that should make comparative studies of the two proteins possible has been eagerly awaited.
Here we report the cloning and characterization of the gene for M. luteus UV endonuclease. Unexpectedly, the deduced primary structure of the protein reveals only a low degree of overall homology to that of the T4 enzyme. We find, however, that not all but many of the amino acid residues implicated in the action of the latter enzyme are conserved in the M. luteus enzyme.
MATERIALS AND METHODS
M. luteus Strains and Culture Conditions.
M. luteus strain ATCC4698 (15) was wild type, and strain DB7, a derivative of ATCC4698 (2), was a double mutant deficient in UV endonuclease and the Uvr homolog (8). Except in transformation experiments (see below), they were cultured at 35°C in nutrient broth [1% tryptone (Difco)/0.5% bonito extract (Wako Pure Chemical, Osaka)/0.5% NaCl] or on nutrient agar (nutrient broth plus 1.5% agar).
UV Irradiation.
Two methods, irradiation with 254-nm UV and acetone-mediated molecular photosensitization (to be referred to simply as photosensitization) were used. The former was done under a germicidal lamp at a dose rate of 1.1 J/m2 per s; the latter was performed by irradiating samples containing 10% (vol/vol) acetone under a 312-nm light source (VL-30M, Vilber Lourmat, Marne La Vallee, France) with a UV-31 filter (Toshiba, Tokyo).
UV Endonuclease Assay.
The reaction mixture (20 μl) contained 10 mM Tris·HCl (pH 7.5), 20 mM EDTA, and appropriate amounts of substrate DNA and the enzyme, and incubation was made for 60 min at 37°C. For the routine assay, the closed circular form of pUC18 (16) was used as substrate. The DNA was given either 60 J/m2 of 254-nm UV or a photosensitization dose of 6 min at a distance of 5.4 cm, and 0.3–0.7 μg of it was used per reaction. After being incubated, the mixture was analyzed by electrophoresis on a 0.8% agarose gel for conversion to open circles. For the analysis of the reaction mode, a 50-bp defined substrate, custom-synthesized by Kurabo (Tokyo), was used. The dithymidylate-containing strand of it (TACACACACGTATGCACATGTTATACGCACACACAGTGCATACACATATA) was labeled with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Toyobo, Tokyo) (17), and annealed with the unlabeled complementary strand. The resulting double-stranded substrate received photosensitization treatment at a distance of 4 cm for 60 min. After being incubated, the reaction containing 90 fmol of the substrate was mixed with an equal volume of the formamide loading solution (17), heated at 95°C for 3 min, and subjected to electrophoresis on a 15% polyacrylamide sequencing gel followed by autoradiography.
Purification of UV Endonuclease.
Preparation of a crude extract of M. luteus ATCC4698 cells and its fractionation were done essentially as described previously up to the phosphocellulose step (15). The phosphocellulose fraction, in 5 mM potassium phosphate buffer (pH 7.0) containing 10% (vol/vol) glycerol, was applied to a DNA cellulose column (1.6 × 4.5 cm, Pharmacia), which was then washed thoroughly with the same buffer and eluted with 40 ml of a 0–1.0 M linear NaCl gradient in the same buffer. Fractions of 0.7 ml each were collected and examined for protein profiles as determined by SDS/PAGE and UV endonuclease activity. Purification of UV endonuclease from E. coli cells was carried out in the same way as above, except that the cells were disrupted by sonication without prior treatment with lysozyme. SDS/PAGE was carried out on a 12.5% gel. Recovery of protein from an SDS/PAGE gel was done as described (18).
Amino-Terminal Sequence Determination.
This was performed in a Model 476A peptide sequencer (Applied Biosystems) by using samples from SDS/PAGE gels.
Construction and Screening of DNA Library.
Size-fractionated BamHI fragments of ATCC4698 DNA (3–15 kbp, 0.3 μg), prepared by agarose gel electrophoresis followed by treatment with QIAEX II (Qiagen, Chatsworth, CA), were ligated to λ EMBL3 BamHI arms (1.0 μg, Stratagene). The resulting DNA was packaged into phage particles with the In vitro packaging kit (Amersham), and amplified in cells of E. coli strain XL1-Blue MRA(P2) (Stratagene). The library was screened by plaque hybridization (17) with an oligonucleotide probe custom-synthesized by Sawady Technology (Tokyo). Labeling and detection of the probe were made with the enhanced chemiluminescence 3′-oligolabeling and detection systems (Amersham).
Plasmid Construction.
This was carried out by standard procedure (17) with pUC18, pT7T3 18U (Pharmacia), and pET-11a (19) as vectors. Construction of the pET-11a derivative involved the conversion of the SphI recognition site to an NdeI site, which was carried out according to the protocol of Hemsley et al. (20). The host for the pUC18- and pT7T3 18U-based plasmids were E. coli strain DH5α (21), whereas that for the pET-11a derivative was strain BL21(DE3) (19).
Nucleotide Sequence Analysis.
This was carried out by the chain termination method (22), with the automated laser fluorescenceDNA sequencer and the Autoread sequencing kit (Pharmacia). Alignment of amino acid sequences was done by the geneworks program (IntelliGenetics).
Complementation in E. coli.
Strain AB2500 (23), an E. coli uvrA6 mutant, was transformed with appropriate recombinant plasmids as described (24). The resulting transformants were grown at 37°C to a mid-logarithmic phase in l-broth [1% tryptone (Difco)/0.5% yeast extract (Difco)/0.5% NaCl], supplemented with ampicillin (50 μg/ml), and the cells were collected by centrifugation. After being washed with phosphate-buffered saline (PBS), the cells were suspended in a 9:1 (vol/vol) mixture of PBS and acetone at twice the original cell density. The cell suspension was left at room temperature for 5 min and then irradiated with 312-nm light. Samples taken at intervals were diluted appropriately in PBS, and spread on l-agar (l-broth plus 1.5% agar) plates supplemented with ampicillin (50 μg/ml), which were then incubated at 37°C. When 254-nm UV was used, the cells were suspended in PBS and irradiated. Strain AB2497 (23), a uvr+ strain that was otherwise isogenic to AB2500, served as control.
In Vitro Gene Expression.
Five micrograms of each pT7T3 18U-based recombinant plasmid was linearized with HindIII, recovered by ethanol precipitation, and subjected to in vitro transcription with the MEGAscrip in vitro transcription kit (Ambion, Austin, TX) according to the manufacturer’s recommendations. Subsequent in vitro translation was performed with one-fifth of the resulting transcripts by use of the rabbit reticulocyte lysate system (Amersham) as recommended by the supplier.
Induced Expression in E. coli.
Cells of strain BL21(DE3) harboring the pET-11a derivative were grown at 37°C in Terrific broth (25), supplemented with ampicillin (100 μg/ml) to a mid-logarithmic phase, isopropyl-1-thio-β-d-galactoside was added to 0.5 mM, and the culture was further incubated for 2 hr.
Transformation of M. luteus.
This was carried out as described (26), with a circular recombinant plasmid serving as transforming DNA. [A circular recombinant plasmid and the linear insert excised therefrom have comparable transforming efficiencies in M. luteus (7).]
RESULTS
Gene Cloning.
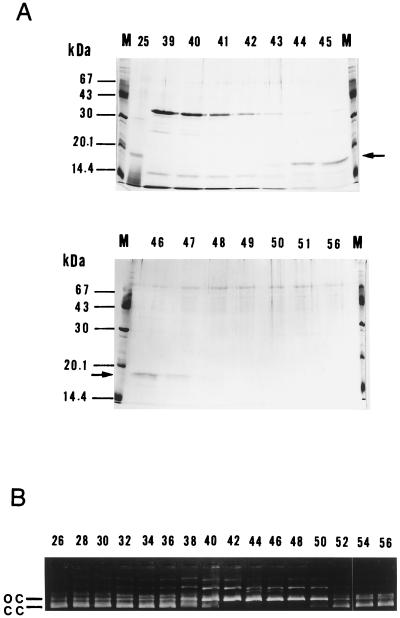
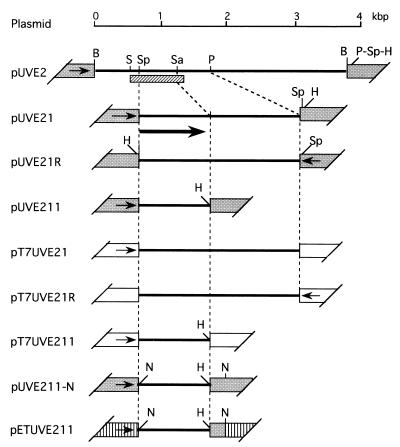
In the DNA cellulose step of purification, the elution profile of UV endonuclease activity was almost the same as that of an 18-kDa protein as revealed by SDS/PAGE (Fig. 1). In addition, we were able to recover the activity by eluting such a band from the gel (data not shown). These results indicated that the 18-kDa protein actually represented UV endonuclease. The amino-terminal sequence of this protein was shown to be MRLWTLH-, and the corresponding oligonucleotide probe UDE1 [ATGCG(AGCT)CT(GCT)TGGAC(GCT)C(GCT)CA] enabled us to clone a 3.8-kbp fragment of M. luteus DNA by screening ≈2 × 104 recombinant phage particles by plaque hybridization. This fragment was recloned into pUC18 to give pUVE2, from which plasmids carrying its UDE1-hybridizable subfragments (pUVE21, pUVE21R, and pUVE211) were derived (Fig. 2).
Figure 1.
Purification of UV endonuclease. M. luteus ATCC4698 cells (≈80 g wet weight) were fractionated as described, and the final DNA cellulose step was monitored for protein profiles and enzyme activity. (A) SDS/PAGE. Aliquots (40 μl) of fractions were analyzed on 12.5% gels, followed by staining with Coomassie blue. Figures, fraction numbers; arrows, the 1.8-kDa band; M, molecular mass standards. (B) UV endonuclease activity. Aliquots (3 μl) of fractions were subjected to UV endonuclease assay using the pUC18 substrate irradiated with 254-nm UV. Figures, fraction numbers; cc, closed circles; oc, open circles.
Figure 2.
Plasmid constructs. pUVE2 (see text) was digested with PstI and self-ligated. The resulting plasmid was cut with SphI, and the 1.1-kbp fragment formed was recloned into pUC18 to give pUVE21 and its reverse-oriented partner pUVE21R. pUVE21 was digested with SacII and HindIII, and the resulting larger fragment was blunt-ended and circularized with a phosphorylated HindIII linker (Takara, Kyoto) to give pUVE211. For the pT7T3 18U-based constructs, the SphI–HindIII fragments were prepared from pUVE21, pUVE21R, and pUVE211, and ligated to pT7T3 18U linearized with SphI and HindIII to generate pT7UVE21, pT7UVE21R, and pT7UVE211, respectively. For construction of pETUVE211, the SphI site of pUVE211 was converted to an NdeI site to make pUVE211-N, and its smaller NdeI fragment was inserted into the NdeI site of pET11a. Solid lines, M. luteus sequences; grey trapezoids, pUC18 sequences (arrows, orientation of lac promoter); open trapezoids, pT7T3 18U sequences (arrows, orientation of T7 promoter); hatched trapezoids, pET11a sequences (arrow, orientation of T7 promoter); hatched bar, sequenced region; thick arrow, the open reading frame identified. Restriction sites: B, BamHI; H, HindIII; N, NdeI; P, PstI; S, SalI; Sa, SacII; Sp, SphI.
Determination of Nucleotide Sequence.
The nucleotide sequence of a region covering the shortest UDE1-hybridizable subfragment (see Fig. 2) was determined (Fig. 3A; GenBank accession no. D78321D78321). The 624-bp region contained an open reading frame (ORF), which encoded a putative protein composed of 154 amino acid residues with a calculated molecular mass of 17,120 Da. The amino-terminal seven residues of the deduced sequence coincided with that of the purified UV endonuclease. A candidate ribosome-binding site was noted 11 to 8 bases upstream of the provisional start codon.
Figure 3.
Sequence analysis of the cloned DNA. (A) Nucleotide sequence with predicted amino acid sequence of the provisional ORF product. Box, tentative ribosome-binding site; overlines, the SphI and SacII cleavage sites (see Fig. 2); underline, the seven N-terminal amino acid residues determined with the purified protein. (B) Comparison of the deduced amino acid sequence of the provisional ORF product with that of T4 endonuclease V (27). The pairs of identical amino acids are marked by asterisks. Upper row, the ORF product; lower row, endonuclease V.
Amino Acid Sequence Similarity to T4 Endonuclease V.
A number of gaps and breaks were required to align the amino acid sequences of the putative ORF product and T4 endonuclease V, and, of the 115 possible amino acid matches only 31 (27%) were identical (Fig. 3B). It was also noted that, of the amino acid residues previously proposed as important in the T4 enzyme (11, 12), Arg-3, Arg-22, Glu-23, and Lys-121 were conserved in the present sequence as Arg-2, Arg-21, Glu-22, and Lys-117, respectively. [Since the amino-terminal methionine of the T4 enzyme is removed in vivo (12), the first three of those conserved residues are situated in exactly the same positions in the two proteins.] However, Thr-2, Arg-26, and Arg-117, also said to be important in the T4 enzyme (11), found no counterparts in the M. luteus sequence. Trp-128 of the T4 enzyme, for which conflicting results had been reported regarding its functional importance (13, 14), was among the conserved residues.
Complementation of E. coli uvrA.
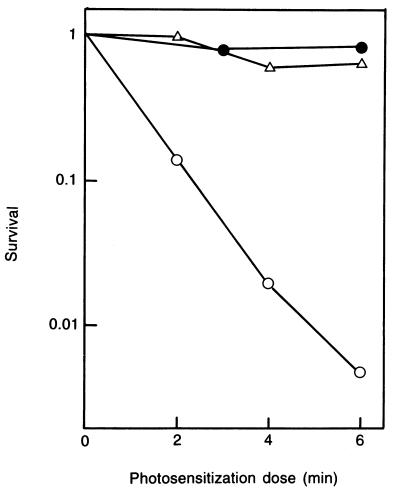
To clarify the identity of the ORF, we determined if the cloned DNA fragment was able to complement the E. coli uvr mutation with respect to the removal of CPyDs from the cellular DNA. It was shown that pUVE211, containing the ORF and little else, was capable of nearly complete suppression of the increased sensitivity of the uvrA cells to photosensitization (Fig. 4). (pUVE211 lacked the original ribosome-binding site for the ORF; presumably, a substitute must have been supplied by the vector.) When irradiated at 254 nm, the cells carrying the plasmid showed partial resistance (data not shown). This was consistent with the stringent specificity of UV endonuclease for CPyDs: photosensitization is known to yield only this class of DNA damage (28), whereas 254-nm UV produces other classes of lethal damage including the (6–4) photoproducts as well as CPyDs.
Figure 4.
Effect of the cloned DNA fragment on cell killing by photosensitization in the E. coli uvrA mutant. Symbols: (•), AB2497 (uvrA+); (○), AB2500 (uvrA6)/pUC18; (Δ), AB2500/pUVE211.
In Vitro Gene Expression.
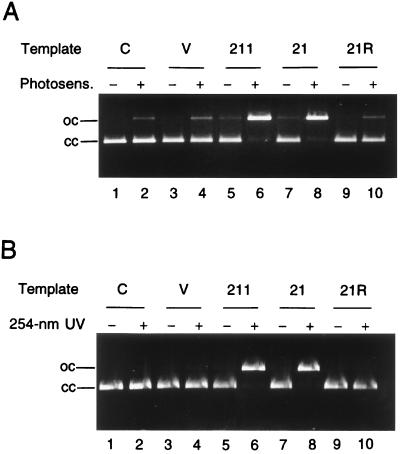
The inserts in pUVE21 and pUVE211 were recloned into pT7T3 18U (Fig. 2), and served as templates for in vitro transcription/translation. It was shown that pT7UVE21 and pT7UV211, but not pT7UVE21R, were capable of generating an activity that nicked CPyD-containing DNA (Fig. 5). These findings lent further support to the notion that the ORF found in the cloned DNA actually encoded UV endonuclease.
Figure 5.
Cleavage of CPyD-containing DNA by the in vitro transcription/translation product. The pUC18 substrate was treated by photosensitization (A) or 254-nm irradiation (B). Two-microliter portions of the translation mixtures (total volume, 50 μl) were used for the assay. Even-numbered lanes, treated substrate; odd-numbered lanes, untreated substrate. Lanes 1 and 2, substrate alone. Transcription templates: pT7T3 18U (lanes 3 and 4), pT7UVE211 (lanes 5 and 6), pT7UVE21 (lanes 7 and 8), and pT7UVE21R (lanes 9 and 10). cc, closed circles; oc, open circles.
Overproduction in E. coli.
An induced expression of the ORF in E. coli cells was attained by the use of pETUVE211 (Fig. 2), in which the gene was under the control of the T7 transcription system that was to be turned on by the lac operon inducer isopropyl-1-thio-β-d-galactoside. Using the same protocol as in M. luteus, we were able to obtain at least 1 mg of purified protein with UV endonuclease activity from about 8 g (wet weight) of induced cells. The identity of the protein was confirmed by showing that its amino-terminal sequence was MRLWTLHPR-.
Confirmation of the Cleavage Mode.
The ability of the ORF product to cleave photosensitization-treated DNA clearly indicated that its activity was specific for CPyDs, but how it cleaved such DNA remained to be seen. We therefore addressed this uncertainty using the protein purified from cells of the E. coli overproducer.
UV endonuclease is known to cleave the N-glycosylic bond between the 5′ pyrimidine moiety in a CPyD and the corresponding deoxyribose portion and to cut the sugar–phosphate backbone on the 3′ side of the resulting abasic deoxyribose by β-elimination (4, 5). Furthermore, the glycosylase reaction has been supposed to proceed via an imino enzyme-substrate intermediate as in T4 endonuclease V, which predicts inhibitions by cyanides and borohydrides previously shown to occur in the T4 enzyme (29).
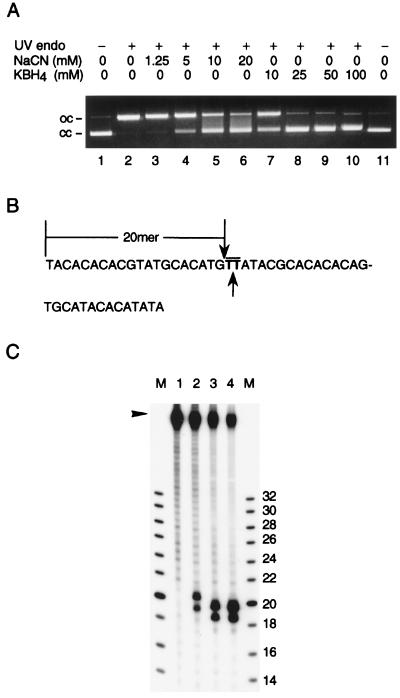
First, we looked at the effects of NaCN and KBH4 on the cleavage of photosensitization-treated DNA. Our results demonstrated that either chemical inhibited the cleavage reaction in a concentration-dependent manner, indicating that the above surmise was actually the case (Fig. 6A). We next determined the site of the backbone scission using the synthetic 50-bp substrate containing a single CPyD (Fig. 6B). The 20-mer fragment produced by the restriction enzyme NlaIII or NspI served as reference. In accordance with the β-elimination mechanism, cleavage by the ORF product occurred between the two thymidine moieties as evidenced by the production of a fragment one “nucleotide” longer than the 20-mer reference fragment (Fig. 6C). [The 19-mer (lanes 3 and 4) and 20-mer (lane 2) cleavage products accompanying the above-mentioned fragments can be accounted for by the 49-mer contaminating the substrate.] From these results, it was firmly established that the ORF product represented a CPyD–DNA glycosylase/abasic lyase or the traditional UV endonuclease.
Figure 6.
Characterization of the DNA cleavage reaction. The cloned-gene product purified from E. coli cells was used. (A) Inhibition by NaCN and KBH4. Each reaction contained 0.3 μg of photosensitization-treated pUC18 DNA, 10 ng of the enzyme, and indicated concentration of the inhibitor. Lanes 1 and 11, substrate alone; lane 2, without inhibitor; lanes 3–6, with NaCN; lanes 7–10, with KBH4. oc, open circles; cc, closed circles. (B) The labeled strand of the 50-bp synthetic substrate. Overline, dithymidylate; downward arrow, cleavage site for NlaIII and NspI; upward arrow, cleavage site for CPyD–DNA glycosylase/abasic lyase. (C) Cleavage of photosensitization-treated (lanes 1 and 2) or untreated (lanes 3 and 4) 50-bp substrate. Enzymes used: none (lane 1), 240 ng of the cloned-gene product (lane 2), 4 units of NlaIII (lane 3), 10 units of NspI (lane 4). M, oligonucleotide size markers (Pharmacia) labeled in the same way as the substrate; figures, chain lengths. In addition to the full-length 50-mer (arrowhead), the substrate preparation contained significant amounts of shorter, mostly 49-mer, contaminants.
Transformation of UV Endonuclease-Defective Mutant.
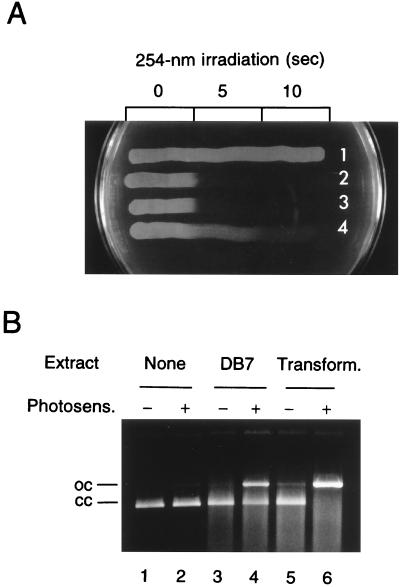
The M. luteus mutant DB7 (2) has been shown to be dually defective in the Uvr homolog as well as in UV endonuclease (8), and more UV sensitive than the single uvr mutant (9). This allowed us to determine if the cloned fragment could restore UV endonuclease to DB7 by transformation. [When the whole chromosomal DNA is used to transform DB7 to UV resistance, uvr+ transformants, which are as resistant to UV as wild type, predominate, making the detection of UV endonuclease-restored transformants virtually impossible (6).] After being incubated with the cloned DNA (the circular form of pUVE211), the cells were plated, irradiated by 254-nm UV for enrichment of transformants (11 J/m2; survival of DB7, ≈10−3), and allowed to form colonies. Six of 15 colonies from one such transformation and two of 10 from another turned out to consist of cells that showed UV resistance intermediate between wild type and DB7 and apparently normal levels of UV endonuclease activity (Fig. 7). Controls without transforming DNA yielded no such colonies (none among 52 and 46 colonies tested, respectively), indicating that reversions or suppressor mutations were too infrequent, if at all, to account for these results. This was an additional proof that the ORF encoded UV endonuclease. Interestingly, the extract from DB7 cells possessed residual nicking activity against CPyD-containing DNA.
Figure 7.
Transformation of DB7. (A) UV sensitivity testing of representative strains. Nutrient broth cultures of strains to be tested were streaked on a nutrient agar plate, which was UV irradiated at 254 nm for indicated time with appropriate masking and incubated. 1, ATCC4698; 2, DB7; 3, DB7 (survivor of the irradiation); 4, putative transformant. (B) UV endonuclease activity of crude extracts. Cells from late-logarithmic phase cultures in nutrient broth (5 ml) were suspended in 0.75 ml of 5 mM potassium phosphate buffer (pH 7.0) containing 10% (vol/vol) glycerol, lyzed by lysozyme treatment (at 1 mg/ml and 37°C for 15 min), and sonicated. After centrifuging the sonicates at 12,000 rpm for 10 min in the cold, 2-μl portions of the supernatants were used for the assay. Even-numbered lanes, photosensitization-treated pUC18 substrate; odd-numbered lanes, untreated substrate. Extracts used: none (lanes 1 and 2), DB7 (lanes 3 and 4), the transformant (lanes 5 and 6). cc, closed circles; oc, open circles.
DISCUSSION
UV endonuclease of M. luteus is characterized by (i) chromatographic behavior typical of a basic protein (adsorbed by phosphocellulose but not by DEAE cellulose under appropriate conditions) (15, 30), (ii) a small molecular mass (<20 kDa; refs. 15 and 30), (iii) absence of inhibition by EDTA (1–3), (iv) a stringent specificity for CPyD-containing DNA (4), (v) combined DNA glycosylase and abasic lyase activities (4, 5), and (vi) deficiency in the mutant DB7 (2, 8). The product of the gene cloned in this study was shown to possess all of these properties. We therefore conclude that we have cloned the gene for UV endonuclease and propose to name it uveA.
Recently, a novel “UV endonuclease” also specific for CPyDs has been described in M. luteus (31). It is a 30,340 Da protein, with an amino acid sequence showing a high degree of homology to that of E. coli endonuclease III, an enzyme specific for thymine glycol and other oxidative products in DNA. We surmise that this enzyme is likely to represent an entity distinct from the traditional UV endonuclease. It may possibly account for the residual nicking activity observed in the extract of DB7 cells (see Fig. 7B). At any rate, this bacterium now seems to have three different enzymes with the potential to initiate the removal of CPyDs from its DNA. To our knowledge, no other organisms have ever been shown to be so heavily armored specifically against this class of DNA damage.
The predicted amino acid sequence of UV endonuclease yields some interesting implications when compared with that of T4 endonuclease V. One implication is concerned with the structure–function relationship for this class of enzymes. Glu-23 and Arg-3 of the T4 enzyme have been shown to be essential for catalysis and substrate binding, respectively (11, 12). These residues, along with two others (Arg-22 and Lys-121) implicated in substrate binding (11), are conserved in UV endonuclease, implying that the two enzymes share a common mechanism for catalysis. It must be noted, however, that although Thr-2, Arg-26, and Arg-117 of the T4 enzyme are reportedly important in substrate binding (11), they are not conserved in the M. luteus enzyme. This may be a reflection of a possible difference between these proteins in the three-dimensional structure; the relatively large gaps required for sequence alignment might also be consistent with this interpretation. A second implication concerns the evolutionary relationship between the two proteins. Their overall sequence similarity was marginal; judging from the large gaps introduced, the observed 27% identity might be an overestimate of the true homology. This is rather unexpected for the proteins with strikingly similar properties, and in sharp contrast to the high amino acid sequence homologies found in Uvr proteins of E. coli and M. luteus (7, 8). An obvious possibility is that the genes of both proteins have diverged from a common ancestor early in the history of evolution. Without the knowledge of the origin of phage T4 it would not be unrealistic to postulate that the hypothetical ancestor was a member of the primordial UV-protective system, which must have come into existence shortly after the advent of life on Earth. Another possibility would be that these proteins might have evolved at unusually high rates after divergence. Sequence data of homologous proteins from other organisms would be essential for addressing this intriguing problem.
Finally, it is anticipated that the E. coli overproduction system for UV endonuclease reported herein will facilitate the study of this protein.
Acknowledgments
This paper is dedicated to the memory of Shunzo Okubo (1930–1978), with whom one of us (H.N.) set out to study UV endonuclease in 1966. We thank Dr. Y. Ito (Kyushu University) for kindly performing the amino-terminal sequence determination for the recombinant protein and Ms. K. Sakai for her supportive services in the laboratory.
Footnotes
References
- 1.Takagi Y, Sekiguchi M, Okubo S, Nakayama H, Shimada K, Yasuda S, Nishimoto T, Yoshihara H. Cold Spring Harbor Symp Quant Biol. 1968;33:219–227. doi: 10.1101/sqb.1968.033.01.025. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan J C, Kushner S R, Grossman L. Proc Natl Acad Sci USA. 1969;63:144–151. doi: 10.1073/pnas.63.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrier W L, Setlow R B. J Bacteriol. 1970;102:178–186. doi: 10.1128/jb.102.1.178-186.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grafstrom R H, Park L, Grossman L. J Biol Chem. 1982;257:13465–13474. [PubMed] [Google Scholar]
- 5.Bailly V, Sente B, Verly W G. Biochem J. 1989;259:751–759. doi: 10.1042/bj2590751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okubo S, Nakayama H, Takagi Y. Biochim Biophys Acta. 1971;228:83–94. [PubMed] [Google Scholar]
- 7.Shiota S, Nakayama H. Mol Gen Genet. 1988;213:21–29. doi: 10.1007/BF00333393. [DOI] [PubMed] [Google Scholar]
- 8.Shiota S, Nakayama H. Mol Gen Genet. 1989;217:332–340. doi: 10.1007/BF02464901. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama H, Shiota S, Umezu K. Mutat Res. 1992;273:43–48. doi: 10.1016/0921-8777(92)90048-8. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 11.Doi T, Recktenwald A, Karaki Y, Kikuchi M, Morikawa K, Ikehara M, Inaoka T, Hori N, Ohtsuka E. Proc Natl Acad Sci USA. 1992;89:9420–9424. doi: 10.1073/pnas.89.20.9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manuel R, Latham K A, Dodson M L, Lloyd R S. J Biol Chem. 1995;270:2652–2661. doi: 10.1074/jbc.270.6.2652. [DOI] [PubMed] [Google Scholar]
- 13.Ishida M, Kanamori Y, Hori N, Inaoka T, Ohtsuka E. Biochemistry. 1990;29:3817–3821. doi: 10.1021/bi00468a002. [DOI] [PubMed] [Google Scholar]
- 14.Latham K A, Carmical J R, Lloyd R S. Biochemistry. 1994;33:9024–9031. doi: 10.1021/bi00196a021. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama H, Okubo S, Takagi Y. Biochim Biophys Acta. 1971;228:67–82. doi: 10.1016/0005-2787(71)90547-8. [DOI] [PubMed] [Google Scholar]
- 16.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Vales L D, Rabin B A, Chase J W. J Biol Chem. 1982;257:8799–8805. [PubMed] [Google Scholar]
- 19.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 20.Hemsley A, Arnheim N, Toney M D, Cortopassi G, Galas D J. Nucleic Acids Res. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmann B. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushner S R. In: Genetic Engineering. Boyer H S, Nicosia S, editors. Amsterdam: Elsevier; 1978. pp. 17–23. [Google Scholar]
- 25.Tartof K D, Hobbs C A. Bethesda Res Lab Focus. 1987;9:12. [Google Scholar]
- 26.Okubo S, Nakayama H. Biochem Biophys Res Commun. 1968;32:825–830. doi: 10.1016/0006-291x(68)90315-x. [DOI] [PubMed] [Google Scholar]
- 27.Valerie K, Henderson E E, deRiel J K. Nucleic Acids Res. 1984;12:8085–8096. doi: 10.1093/nar/12.21.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Ishai R, Ben-Hur E, Hornfeld Y. Isr J Chem. 1968;6:769–775. [Google Scholar]
- 29.Dodson M L, Schrock R D, III, Lloyd R S. Biochemistry. 1993;32:8284–8290. doi: 10.1021/bi00083a032. [DOI] [PubMed] [Google Scholar]
- 30.Riazuddin S, Grossman L. J Biol Chem. 1977;252:6280–6286. [PubMed] [Google Scholar]
- 31.Piersen C E, Prince M A, Augustine M L, Dodson M L, Lloyd R S. J Biol Chem. 1995;270:23475–23484. doi: 10.1074/jbc.270.40.23475. [DOI] [PubMed] [Google Scholar]