Abstract
The biosynthesis of small molecules can be fine-tuned by (re)engineering metabolic flux within cells. We have adapted this approach to optimize an in vivo selection system for the conversion of prephenate to phenylpyruvate, a key step in the production of the essential aromatic amino acid phenylalanine. Careful control of prephenate concentration in a bacterial host lacking prephenate dehydratase, achieved through provision of a regulable enzyme that diverts it down a parallel biosynthetic pathway, provides the means to systematically increase selection pressure on replacements of the missing catalyst. Successful differentiation of dehydratases whose activities vary over a >50,000-fold range and the isolation of mechanistically informative prephenate dehydratase variants from large protein libraries illustrate the potential of the engineered selection strain for characterizing and evolving enzymes. Our approach complements other common methods for adjusting selection pressure and should be generally applicable to any selection system that is based on the conversion of an endogenous metabolite.
Keywords: directed evolution, enzyme design, genetic complementation, prephenate dehydratase, tetracycline promoter
Evolutionary strategies that couple random mutagenesis with high-throughput screening or selection have been successfully exploited to create and characterize protein catalysts on a human time scale (1–6). Such approaches afford insight into the complex interactions that influence protein folding, structure, and catalytic mechanism. They can also aid in the design of new proteins with tailored catalytic activities and selectivities.
Selection methods are particularly powerful in this context because they allow exhaustive analysis of protein libraries containing >107 members. Many in vivo selection schemes rely on the capability of a protein to complement a metabolic defect in a microorganism (3). Such strains can be engineered by mutating or deleting an endogenous gene that encodes an enzyme with the desired activity. Designing this type of selection system is nontrivial, however. Living organisms are complex, and unanticipated mechanisms may compensate for the engineered defect. As a consequence, it is generally difficult to predict the threshold of activity needed for survival of an auxotrophic host and the dynamic range of selectable activities.
The ideal selection system should also allow adjustment of selection pressure to the task at hand. For example, discovery of active variants in large protein libraries may initially require a low selection hurdle, whereas increasing stringency is required for their evolutionary optimization in subsequent selection rounds. Because selection pressure is linked to intracellular catalyst concentration, stringency can be adjusted in practice by altering gene dose or promoter strength or by targeting the encoded catalyst for degradation (7, 8). Selection pressure is also sensitive to the concentration of critical metabolites and can sometimes be varied simply by changing amounts of substrate added to the growth medium (7). In vivo, resource allocation is often accomplished by controlling the flux of branch point intermediates in metabolic networks (9). We wondered whether an analogous approach might be used to fine-tune the stringency of genetic selection systems involving transformations of substrates that are produced biosynthetically by the host.
The shikimate biosynthetic pathway (10), having multiple branch points, is a good system to evaluate strategies for tuning selection pressure. In this study we focus on prephenate dehydratase (PDT), which catalyzes the conversion of prephenate to phenylpyruvate, the penultimate step in the biosynthesis of l-phenylalanine (Fig. 1). In the cell, prephenate is also converted by prephenate dehydrogenase (PDH) to 4-hydroxyphenylpyruvate, the immediate precursor to l-tyrosine. Negative feedback inhibition of the dehydrogenase by tyrosine helps to regulate the relative flux down the two branches of the pathway (11, 12).
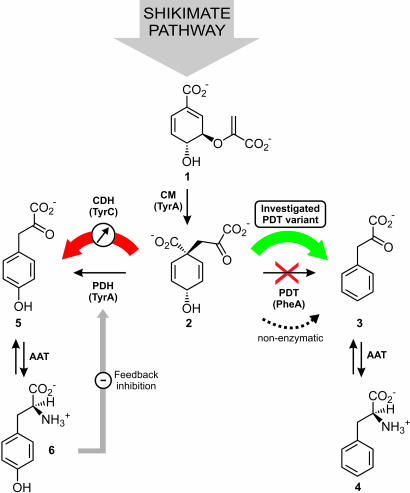
Fig. 1.
The engineered PDT selection system. The red X symbolizes the absence of the chorismate mutase–PDT (PheA) in the E. coli KA34 host strain. As a result of the pheA gene deletion, prephenate (2), generated from chorismate (1) by chorismate mutase (TyrA), is converted enzymatically to l-tyrosine (6) via 4-hydroxyphenylpyruvate (5) by prephenate dehydrogenase (TyrA) and AAT (aromatic amino acid aminotransferase). However, prephenate can accumulate because of feedback inhibition of TyrA by l-tyrosine. Spontaneous nonenzymatic conversion of prephenate to phenylpyruvate (3), which is subsequently transformed to l-phenylalanine (4) by AAT, leads to “leaky” growth under selective conditions. By introducing an externally regulable CDH (TyrC) from Z. mobilis, the intracellular prephenate concentration can be reduced, diminishing background growth in the absence of exogenously added phenylalanine and also affording tunable selection pressure on dehydratase surrogates.
Here we show that selection stringency of a phenylalanine auxotroph lacking an endogenous dehydratase can be systematically adjusted by regulating the production of a second enzyme that diverts prephenate away from the phenylalanine branch. Controlling the intracellular concentration of prephenate in this way eliminates problems associated with background growth due to its spontaneous conversion to phenylpyruvate. At the same time, rapid sorting and isolation of dehydratase catalysts according to activity over a >50,000-fold range is facilitated.
Results
Engineering the PDT Selection System.
To select for PDT activity, a bacterial strain lacking the corresponding natural enzyme is needed. We used a new homologous recombination method (13) to delete the pheA gene, which encodes a bifunctional chorismate mutase–PDT, from the genome of a RecA-deficient Escherichia coli strain. Because of the resulting defect in the shikimate biosynthetic pathway, the KA34 strain shows severely compromised growth unless the essential amino acid phenylalanine or a functional dehydratase is provided. Nevertheless, the auxotroph does grow slowly under some selective conditions. Specifically, when the cells are plated at high density (>107 cells per 30-ml agar plate), >30 μg/ml tyrosine is added to the medium, the intracellular chorismate mutase concentration is increased, or the pH of the medium is <6.0, background growth is sufficiently high to complicate selection experiments.
Because phenylalanine auxotrophs are known to accumulate prephenate (14), residual growth under selective conditions may be due to the relatively facile spontaneous reaction of prephenate to phenylpyruvate. If so, reduction of intracellular prephenate concentration should help to circumvent this problem. We exploited a cyclohexadienyl dehydrogenase (CDH) from Zymomonas mobilis (15) to convert prephenate irreversibly to 4-hydroxyphenylpyruvate rather than to phenylpyruvate (Fig. 1). In contrast to the endogenous prephenate dehydrogenase (TyrA), CDH is not feedback-inhibited by tyrosine.
To enable fine-tuning of the selection system, we placed the CDH-encoding tyrC gene on the helper plasmid pMHEtetCDH (Fig. 2) under the control of a regulable tetracycline-dependent promoter element (tetR-Ptet) (8). The latter affords homogeneous gene expression over a broad dynamic range. In the absence of tetracycline, no CDH is produced and background growth is observed after 10–12 days under selective conditions (i.e., minus phenylalanine). Under these conditions weakly active dehydratases might restore prototrophy, but a large number of clones would have to be examined to find them among the artifacts. Addition of tetracycline to the medium leads to the production of CDH, which reduces the intracellular concentration of prephenate and hence background growth. At tetracycline concentrations >0.4 μg/ml background growth is never detected. In the presence of inducer, complementation of phenylalanine auxotrophy should also require a more efficient PDT, because this enzyme must compete with CDH for the same cellular resource. If higher CDH concentrations necessitate the production of more efficient dehydratases, adjusting the tetracycline concentration should also provide a simple mechanism for regulating selection stringency.
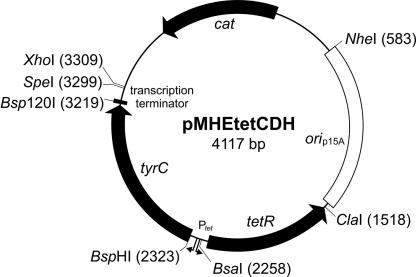
Fig. 2.
Helper plasmid pMHEtetCDH. The plasmid contains a chloramphenicol resistance gene (cat), a p15A origin of replication (orip15A) compatible with the ColE1-type origin of the library plasmid (31), and the CDH gene from Z. mobilis (tyrC), whose transcription is regulated by a tetracycline-dependent promoter element (tetR-Ptet). Unique restriction sites are shown with their position in parentheses.
Calibration of the PDT Selection System.
To assess the dynamic range of the system, KA34/pMHEtetCDH cells were transformed with plasmids encoding dehydratases covering a broad span of activity (Table 1). The transformants were streaked out onto minimal medium plates containing different amounts of tetracycline. As summarized in Table 2, the appearance of single colonies depends on the inducer concentration and the efficiency of the encoded catalyst. Below 0.5 μg/ml tetracycline, a dehydratase with a kcat/Km value as low as 24 M−1·s−1 confers a modest growth advantage over background and, at the same time, can be clearly distinguished from more active catalysts. At intermediate tetracycline concentrations, such weakly active catalysts complement poorly, but moderately active (kcat/Km ≈ 103 M−1·s−1) and highly active (kcat/Km > 104 M−1·s−1) enzymes can be differentiated based on the distinct growth rates they afford. At the highest concentration of tetracycline tested (5 μg/ml), it is even possible to single out the most active catalysts. Thus, by varying the tetracycline concentration, the stringency of the PDT selection system can be systematically tuned to identify dehydratase catalysts spanning a >50,000-fold range in activity.
Table 1.
Kinetic parameters of PDT variants
Table 2.
Calibration of the selection system
| Harbored enzyme variant | Tetracycline concentration, μg/ml |
||||
|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2.5 | 5 | |
| MjPDT | 2 | 2 | 2 | 3 | 3 |
| WT EcPDT | 2 | 2 | 2 | 3 | 5 |
| W226A EcPDT | 2 | 3 | 4 | 7 | >12 |
| N160A EcPDT | 8 | 12 | >12 | >12 | N.G. |
Number of days needed for single-colony formation (colony diameter: 0.5–1 mm) at 30°C. KA34/pMHEtetCDH transformants producing one of the four PDTs were picked from phenylalanine-supplemented plates and streaked out onto selective M9s plates as described in Materials and Methods. N.G., no growth observable.
Model selection experiments confirmed the utility of this system for isolating weakly active enzymes. KA34/pMHEtetCDH was transformed with 1:1, 1:10, and 1:100 mixtures of plasmids encoding either the N160A E. coli PDT (EcPDT) variant or an E. coli chorismate mutase that lacks PDT activity. The ratio of colonies obtained under selective conditions in the absence of phenylalanine relative to the number of colonies on plates containing phenylalanine corresponds well to the ratio of the two transformed plasmids (Table 3).
Table 3.
Model selection experiments for PDT genes
| Ratio of transformed plasmids* (N160A EcPDT:EcCM genes) | Colonies on nonselective plates (M9s + 20 μg/ml l-phenylalanine) | Colonies on selective plates† (M9s + 80 ng/ml tetracycline) | Experimentally derived fraction of complementing clones |
|---|---|---|---|
| 1:1 | 1,900 | 738 | 1:1.3 |
| 1:10 | 1,800 | 158 | 1:11 |
| 1:100 | 800 | 6 | 1:133 |
*Plasmids pAKZ7-N160A (encoding N160A EcPDT; see footnote § in Table 1) and pAKZ3-EcCM (encoding the chorismate mutase domain from E. coli PheA) were mixed in different ratios and introduced into KA34/pMHEtetCDH cells. The model selection experiments were performed as described in Materials and Methods at a tetracycline concentration of 80 ng/ml.
†The identity of representative isolated plasmids from selected colonies was confirmed by restriction analysis.
Application of the PDT Selection System.
Although PDTs from a number of organisms have been biochemically characterized (see for example refs. 16–20), their mechanism of action remains unclear. We have exploited our selection system to investigate two sets of highly conserved residues in this enzyme family. Specifically, using the monofunctional and thermostable PDT from Methanocaldococcus jannaschii (MjPDT) as a template (16), we targeted the tripeptide sequence Thr-172–Arg-173–Phe-174 (TRF) and, independently, three tyrosines (YYY: Tyr-7, Tyr-14, and Tyr-114) for combinatorial mutagenesis. The TRF tripeptide is found in >95% of all PDTs. Based on site-directed mutagenesis experiments with the homologous E. coli enzyme (19), Thr-172 was expected to be important for catalysis. Furthermore, Arg-173, as the only conserved cationic group in the enzyme, seemed like a likely candidate for binding the dianionic substrate, whereas Phe-174, the most highly conserved hydrophobic residue in the PDT family, could constitute part of the active site or play a structural role (16, 19). Positions 7, 14, and 114, which are >85% conserved as phenylalanine, tyrosine, or tryptophan across the entire PDT family, were chosen to test the possibility that the cationic intermediate that forms upon decarboxylation of prephenate (21) might be stabilized by cation–π interactions (22).
The TRF and YYY libraries were constructed from randomized oligonucleotides containing three NNS codons each. After transformation of the KA34/pMHEtetCDH strain, they contained 6.5 × 105 and 6.1 × 105 members, respectively, ensuring adequate coverage of the 3.3 × 104 theoretically possible codon variants.† Both libraries were plated under selective conditions in the absence of phenylalanine using tetracycline to adjust the selection stringency, and plasmids from functional clones were isolated and sequenced to assess the preferences at the targeted sites.
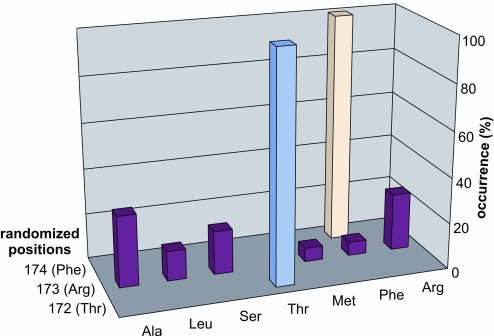
The TRF library was subjected to stringent selection conditions (5 μg/ml tetracycline). Under these conditions, 0.1% of the library was found to complement the dehydratase deficiency. Functional clones contained only WT threonine and phenylalanine residues at positions 172 and 174, whereas Arg-173 was found to be surprisingly tolerant to substitution (Fig. 3).
Fig. 3.
Selection results for the MjPDT TRF library. Amino acid frequencies at the randomized sites were obtained by analysis of 24 functional MjPDT variants, which were isolated under stringent selection conditions (5 μg/ml tetracycline). The WT MjPDT gene was not recovered in this experiment, judging from the observed codons. Under nonselective conditions, the observed amino acid distribution at each position in the tripeptide motif was random. The WT residue at each of the randomized positions is indicated in parentheses.
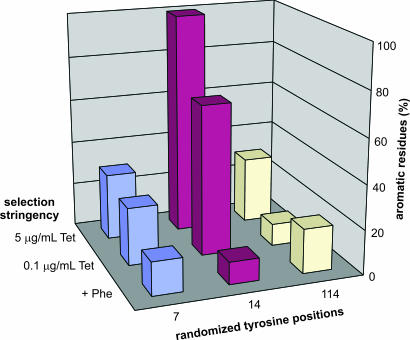
The YYY library was plated on selective medium containing either 0.1 or 5 μg/ml tetracycline. The complementation frequency under the least stringent conditions was ≈15% and decreased to 7% at the higher tetracycline concentration. The occurrence of aromatic amino acids (including histidine) in functional clones obtained under the different selection conditions is shown in Fig. 4. On plates supplemented with phenylalanine, no preference for any amino acid is evident at any of the randomized sites, and the four aromatic residues appear with a frequency that matches expectations for a random distribution (≈13%). Upon selection, aromatics become significantly enriched at position 14, even under the least selective conditions, and at 5 μg/ml tetracycline only aromatic amino acids are found at this site (65% phenylalanine, 15% tyrosine, 15% tryptophan, and 5% histidine). In contrast, only a modest preference for aromatics is observed at positions 7 and 114, suggesting that these residues are not directly involved in catalysis.
Fig. 4.
Frequencies of aromatic amino acids (including His) in MjPDT variants selected from the YYY library at different selection stringencies. The data are based on an analysis of 20 of the largest colonies picked from each plate type. In the presence of phenylalanine, no significant preference for aromatic amino acids is seen at any of the randomized positions. In contrast, under selective conditions, aromatic residues become enriched at position 14 as the selection stringency is increased upon raising the tetracycline concentration.
Discussion
Deleting a key biosynthetic enzyme can severely perturb metabolite flux within a cell, especially under selective conditions. For example, the flow of precursors into the disrupted pathway often results in accumulation of one or more metabolic intermediates upstream of the blocked step (9). Such a situation is potentially problematic for selection experiments, because elevated concentrations of the substrate for the missing enzyme boost the nonenzymatic background reaction and favor isolation of enzyme variants with low substrate affinity.
Our results show that such problems can be minimized or even eliminated through metabolic engineering. Excess substrate can be efficiently removed from cellular metabolism by providing a second enzyme to channel it away from the blocked step. As a consequence, background growth is reduced and greater demands are placed on the enzyme being selected. In our system, regulating the production of CDH, which competes with PDT for prephenate, allows the selection stringency for dehydratase activity to be adjusted over a broad dynamic range. Differentiation of PDTs with as little as a 10-fold difference in kcat/Km over a >50,000-fold range establishes the efficacy of this approach. Efficient isolation of a weakly active dehydratase (kcat/Km = 24 M−1·s−1) further demonstrates the method's utility for directed evolution applications.
Genetic selection is a powerful tool for characterizing enzyme mechanism (3), and we have exploited the PDT selection system to gain insight into MjPDT. Sequence analysis of several hundred PDTs had shown that residues 172–174 in this enzyme are highly conserved across the entire protein family (16), signifying potentially important roles in catalysis. The strict conservation of Thr-172 and Phe-174 in our selection experiments is consistent with this interpretation, but the tolerance of Arg-173 to mutation was unexpected. The crystal structure of a PDT from Staphylococcus aureus Mu50, which was deposited (Protein Data Bank ID code 2IQ8) after completion of this study, provides a rationalization of these results: both Thr-168 and Phe-170 (which correspond to Thr-172 and Phe-174 in MjPDT) point into a cavity that likely constitutes the active site, whereas Arg-169 (Arg-173 in MjPDT) is located on the protein surface (Fig. 5). As previously suggested (19), Thr-172 may thus provide a hydrogen bond to prephenate during catalysis. Phe-174, which is physically adjacent to Thr-172, could also bind the substrate through van der Waals interactions. Although Arg-173 may contribute to protein stability through salt bridges with two surface glutamates (Fig. 5B), the selection and structural data convincingly show that it is not involved in either substrate recognition or catalysis.
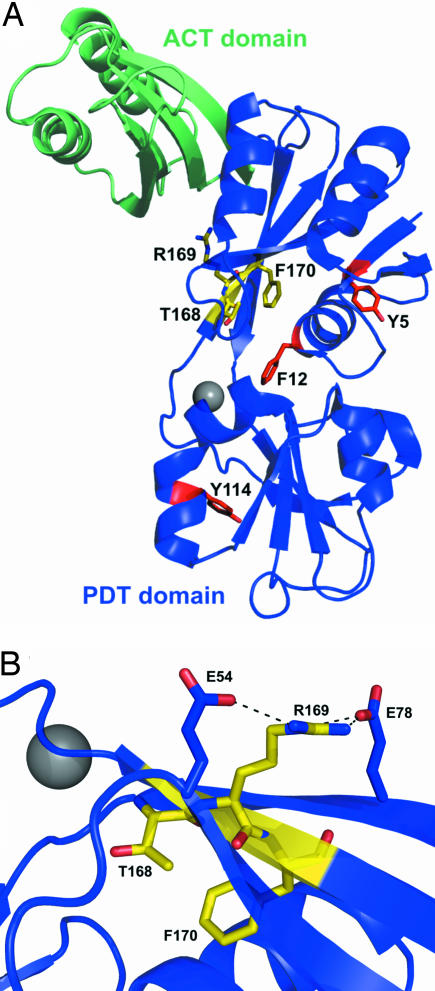
Fig. 5.
Structure of the S. aureus Mu50 PDT (Protein Data Bank ID code 2IQ8). (A) Ribbon diagram showing the PDT domain (blue) and an appended ACT regulatory domain (green). The tripeptide motif randomized in the TRF library is highlighted in yellow, and the aromatic residues randomized in the YYY library are shown in red. The gray sphere symbolizes a bound magnesium atom. The side chains of residues Phe-12, Thr-168, and Phe-170 (corresponding to Tyr-14, Thr-172, and Phe-174 in MjPDT) point into a cavity that probably corresponds to the active site. (B) Close-up view of the TRF tripeptide. Whereas Thr-168 and Phe-170 are directed toward the interior of the protein and line a well defined pocket, Arg-169 (which corresponds to MjPDT Arg-173) is located on the exterior surface, where it engages in salt bridges with Glu-54 and Glu-78.
The selection experiments with the YYY library illustrate how selection stringency can be easily adjusted to probe conservation patterns simply by altering the amount of tetracycline added to the selection plates. Of the three randomized tyrosines, a clear preference for aromatic residues is evident only at position 14, and this preference increases with increasing selection pressure. Although aromatic amino acids at this position are not essential for function, they apparently contribute significantly to the high efficiency required under stringent selection conditions, consistent with the observation that this residue (corresponding to Phe-12 in the Staphylococcus structure) is the only one located in the putative active site (Fig. 5A). Its aromatic side chain could conceivably stabilize the cationic intermediate (21) that is generated upon decarboxylation of prephenate through cation–π interactions (22), but a structural role may be more plausible, given the observed bias for phenylalanine over tryptophan and tyrosine, which are normally preferred at cation–π binding sites, and the appearance of histidine, which is never used for this purpose (23). Structures of the enzyme complexed with substrate or transition state analogs will be needed to clarify the precise nature of this interaction.
Tunable stringency can greatly facilitate efforts to characterize and evolve enzymes by combinatorial mutagenesis and selection. Metabolic regulation of intracellular substrate concentration represents one viable strategy for achieving such control. As shown here, this can be accomplished by diverting the substrate of the target reaction down an alternative biosynthetic pathway using a competing enzyme produced under the control of a tightly regulated promoter. Our approach is not limited to manipulations directly at the selection step, however, because forks are abundant in cellular metabolism. In principle, branching steps earlier in the biosynthetic reaction sequence, or even after the target reaction, could be exploited to modulate metabolic flux and, hence, selection pressure. Although similar control might also be achieved by direct regulation of one of the catalysts needed to biosynthesize the required substrate, using a branch point enzyme to divert flux has the advantage of not requiring additional chromosomal modifications of the host organism. The optimal strategy will necessarily depend on the connectivity of the pathway in question, but an arsenal of established engineering strategies (24) should allow the creation of a regulable selection regime for virtually any metabolic reaction.
Materials and Methods
DNA Manipulations.
Molecular cloning was performed according to standard procedures (25). Restriction endonucleases, Taq polymerase, and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Oligonucleotides were obtained by custom synthesis from Microsynth (Balgach, Switzerland). PCRs were performed using PfuTurbo polymerase from Stratagene (La Jolla, CA) or Taq polymerase. All portions of the constructed plasmids generated by PCR were confirmed by DNA sequencing on a 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA) by chain termination chemistry (26) using the BigDye Terminator Cycle Sequencing Kit from the same company.
Plasmid Constructions.
Details on the construction of plasmids pDpheA, pMHEtetCDH, pAKZ3-EcCM, pAKZ6, pAKZ7, pAKZ7-W226A, pAKZ7-N160A, and pAKZ13 are provided in supporting information (SI) Text.
Construction of the E. coli Phenylalanine Auxotroph KA34.
The PDT-deficient E. coli strain KA34 was derived from KA10, which was constructed from strain YMC9 (27) by generalized P1 transduction to delete the recA gene as described previously (28). The genotype of KA10 is Δ(srlR-recA)306::Tn10, thi-1, endA-1, hsdR17, Δ(argF-lac)U169, supE44. KA34 [genotype KA10 Δ(pheA)::neo] was engineered by using a chromosomal gene targeting technique (13) and the knockout plasmid pDpheA. The chromosomal lesion in KA34 was verified by PCR using different combinations of primers that hybridize to the inserted kanamycin resistance gene and to the chromosomal DNA outside of the segments cloned in pDpheA, followed by restriction analysis of the PCR products. In addition, a plasmid cointegrate was ruled out by testing for ampicillin and UV sensitivity as described previously (13).
PDT Selection System.
KA34/pMHEtetCDH cells additionally transformed with a library plasmid were subjected to selection on M9s agar plates. The latter are based on M9c minimal agar plates (29) but contain 150 μg/ml sodium ampicillin rather than 50 μg/ml, as well as 5 μg/ml l-tyrosine, 0.2 mM isopropyl 1-thio-β-d-galactopyranoside, and 10 μM sodium salicylate. Tetracycline was plated just before use. For nonselective control conditions, M9s was supplemented with 20 μg/ml l-phenylalanine.
Tetracycline Titration and Model Selections.
Each M9s plate containing a given amount of tetracycline had four streak-outs of four different, single colony-purified transformants of KA34/pMHEtetCDH (producing MjPDT, WT EcPDT, W226A EcPDT, or N160A EcPDT) on physically separated agar sectors (prepared by cutting out slices to prevent diffusion of released metabolites). MjPDT was produced from plasmid pAKZ11 (16), WT EcPDT from pAKZ7, and the W226A and N160A variants of EcPDT from plasmids pAKZ7-W226A and pAKZ7-N160A, respectively. The tetracycline titration assay was carried out in duplicate by incubating four identical plates at 30°C at each tetracycline concentration.
For the model selections, plasmids pAKZ7-N160A (encoding N160A EcPDT) and pAKZ3-EcCM (encoding the chorismate mutase domain of E. coli PheA) were mixed in a 1:1, 1:10, and 1:100 ratio. Electrocompetent KA34/pMHEtetCDH cells (50 μl) were transformed with 1 ng of the plasmid mixtures. Transformation, washing, plating, and incubation of the cells were performed as described for the in vivo selection experiments with the libraries (see below), except that only 1/60th of the cells was plated on M9s plates containing 80 ng/ml tetracycline. Plasmids isolated from selected colonies were identified by restriction analysis.
Library Construction and in Vivo Selection.
All randomized library gene fragments were ligated with the 4,520-bp BsaI–BsaI fragment of the library vector pAKZ6 (5,899 bp) (containing a stuffer fragment flanked by two nonpalindromic BsaI restriction sites). The randomized library genes for the TRF library were created by standard overlap extension PCR (30) of library fragments sPCR1 and sPCR2 using the flanking primers pAKZ4_ins_rev2 and MjPDT_cr_lib_C1 for amplification (see below). sPCR1 was prepared with primers pAKZ4_ins_rev2 (5′-GTAGGACGGGTCTCTGAACTTTAAGAAGGAGATATACATATG; BsaI restriction site underlined) and MjPDT_cr_Lib_TRF− (5′-CTTATTATTTTTATAATCTTCAATATTTTCATCC), and sPCR2 was prepared with primers MjPDT_cr_Lib_TRF+ (5′-GGATGAAAATATTGAAGATTATAAAAATAATAAGNNSNNSNNSATTTTAATTGGTAAAAAAGTTAAATTTAAATATCATCC; randomized nucleotides indicated in bold; N is an equimolar mixture of all nucleotides and S indicates a mixture of C and G) and MjPDT_cr_Lib_C1 (5′-GTAGGACGGGTCTCAGTGGTTATTAATCAAAAACTGGGTATTTTCCTAAAAG; BsaI restriction site underlined) using pAKZ13 (linearized with EcoRV, dephosphorylated, and gel-purified) as template.
The YYY library genes were constructed by assembling three overlapping PCR products (sPCR3, sPCR4, and sPCR5), two of which contain the randomized codons, and PCR-amplifying the assembled gene using the flanking primers pAKZ4_ins_rev2 and MjPDT_cr_Lib_C1. Employing pAKZ13 (linearized with EcoRV, dephosphorylated, and gel-purified) as template, sPCR3 was produced by using primers pAKZ4_ins_rev2 and MjPDT_crLib_Y7/Y14− (5′-AATAACTGCTTTATTCATGGATCCAG), sPCR4 was produced by using primers MjPDT_crLib_Y7/Y14+ (5′-CTGGATCCATGAATAAAGCAGTTATTNNSACATTACCAAAAGGAACGNNSAGTGAAAAAGCTACAAAGAAATTTTTAGACTAC; randomized codons in bold) and MjPDT_crLib_Y114− (5′-ATTTCTACATTGAGCTAATGCCTGC), and sPCR5 was produced by using primers MjPDT_crLib_Y114+ (5′-GCAGGCATTAGCTCAATGTAGAAATNNSATAAAAAAGCACGGTTGGGATG; randomized codons in bold) and MjPDT_cr_Lib_C1. These three PCR products were assembled by using the following temperature program: four cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 10 min (the long elongation times diminished the formation of undesired assembly products).
Electrocompetent cells of the KA34 selection strain containing the pMHEtetCDH helper plasmid (50 μl) were transformed with the appropriate plasmid or ligation mixture. Recovery after electroporation was performed for 50 min at 30°C in 18 ml of SOC medium (the large volume of SOC, yielding an OD600 of 0.6, differs from standard protocols and results in an up to 5-fold-higher transformation efficiency of this strain). The cells were washed three times at 4°C with 4 ml of M9 salts (25) to remove residual l-phenylalanine, resuspended in 3 ml of M9 salts, plated in 50-μl portions per M9s plate (containing a defined tetracycline concentration), and incubated at 30°C.
Supplementary Material
Acknowledgments
We are grateful to Prof. Uwe Sauer (Institute of Molecular Systems Biology, ETH Zurich) for chromosomal DNA from Z. mobilis and to Dr. Keith Backman (Massachusetts Institute of Technology, Cambridge, MA) for plasmid pKB663. This work was supported by the Swiss National Foundation. M.H.E. was the recipient of a Marie Curie Intra-European Fellowship.
Abbreviations
- CDH
cyclohexadienyl dehydrogenase
- PDT
prephenate dehydratase
- EcPDT
E. coli PDT
- MjPDT
M. jannaschii PDT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705379104/DC1.
Sequencing of randomly picked clones from nonselective plates from each library showed that approximately half contained one or more additional mutations outside of the intentionally randomized regions because of oligonucleotide synthesis errors or polymerase mistakes during PCR. Nevertheless, no obvious differences in selection preferences were observed between functional variants with and without the unprogrammed mutations, suggesting that they do not influence the observed trends.
References
- 1.Farinas ET, Bulter T, Arnold FH. Curr Opin Biotechnol. 2001;12:545–551. doi: 10.1016/s0958-1669(01)00261-0. [DOI] [PubMed] [Google Scholar]
- 2.Powell KA, Ramer SW, del Cardayre SB, Stemmer WPC, Tobin MB, Longchamp PF, Huisman GW. Angew Chem Int Ed Engl. 2001;40:3948–3959. doi: 10.1002/1521-3773(20011105)40:21<3948::aid-anie3948>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SV, Kast P, Hilvert D. Angew Chem Int Ed Engl. 2001;40:3311–3335. doi: 10.1002/1521-3773(20010917)40:18<3310::aid-anie3310>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 4.Kazlauskas RJ. Curr Opin Chem Biol. 2005;9:195–201. doi: 10.1016/j.cbpa.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Woycechowsky KJ, Vamvaca K, Hilvert D. Adv Enzymol Relat Areas Mol Biol. 2007;75:241–294. doi: 10.1002/9780471224464.ch4. [DOI] [PubMed] [Google Scholar]
- 6.Jaeger K-E, Eggert T, Eipper A, Reetz MT. Appl Microbiol Biotechnol. 2001;55:519–530. doi: 10.1007/s002530100643. [DOI] [PubMed] [Google Scholar]
- 7.Yano T, Oue S, Kagamiyama H. Proc Natl Acad Sci USA. 1998;95:5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neuenschwander M, Butz M, Heintz C, Kast P, Hilvert D. Nat Biotechnol. 2007 doi: 10.1038/nbt1341. in press. [DOI] [PubMed] [Google Scholar]
- 9.Stephanopoulos G, Vallino JJ. Science. 1991;252:1675–1681. doi: 10.1126/science.1904627. [DOI] [PubMed] [Google Scholar]
- 10.Haslam E. Shikimic Acid: Metabolism and Metabolites. New York: Wiley; 1993. [Google Scholar]
- 11.Christopherson RI. Arch Biochem Biophys. 1985;240:646–654. doi: 10.1016/0003-9861(85)90072-4. [DOI] [PubMed] [Google Scholar]
- 12.Turnbull J, Morrison JF, Cleland WW. Biochemistry. 1991;30:7783–7788. doi: 10.1021/bi00245a017. [DOI] [PubMed] [Google Scholar]
- 13.Gamper M, Kast P. Biotechniques. 2005;38:405–408. doi: 10.2144/05383ST03. [DOI] [PubMed] [Google Scholar]
- 14.Davis BD. Science. 1953;118:251–252. doi: 10.1126/science.118.3061.251. [DOI] [PubMed] [Google Scholar]
- 15.Zhao G, Xia T, Ingram LO, Jensen RA. Eur J Biochem. 1993;212:157–165. doi: 10.1111/j.1432-1033.1993.tb17646.x. [DOI] [PubMed] [Google Scholar]
- 16.Kleeb AC, Kast P, Hilvert D. Biochemistry. 2006;45:14101–14110. doi: 10.1021/bi061274n. and erratum (2007) 46:2552–2552. [DOI] [PubMed] [Google Scholar]
- 17.Fischer R, Jensen RA. Methods Enzymol. 1987;142:507–512. doi: 10.1016/s0076-6879(87)42063-6. [DOI] [PubMed] [Google Scholar]
- 18.Davidson BE. Methods Enzymol. 1987;142:432–439. doi: 10.1016/s0076-6879(87)42054-5. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Wilson DB, Ganem B. Biochemistry. 2000;39:4722–4728. doi: 10.1021/bi9926680. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Williams DS, Aldrich HC, Xie G, Gabriel DW, Jensen RA. Microb Comp Genomics. 1997;2:141–158. doi: 10.1089/omi.1.1997.2.141. [DOI] [PubMed] [Google Scholar]
- 21.Hermes JD, Tipton PA, Fisher MA, O'Leary MH, Morrison JF, Cleland WW. Biochemistry. 1984;23:6263–6275. doi: 10.1021/bi00320a057. [DOI] [PubMed] [Google Scholar]
- 22.Dougherty DA. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 23.Mecozzi S, West AP, Jr, Dougherty DA. Proc Natl Acad Sci USA. 1996;93:10566–10571. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey JE. Science. 1991;252:1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson AR. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backman K, Chen Y-M, Magasanik B. Proc Natl Acad Sci USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kast P, Hennecke H. J Mol Biol. 1991;222:99–124. doi: 10.1016/0022-2836(91)90740-w. [DOI] [PubMed] [Google Scholar]
- 29.Gamper M, Hilvert D, Kast P. Biochemistry. 2000;39:14087–14094. doi: 10.1021/bi0016570. [DOI] [PubMed] [Google Scholar]
- 30.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 31.Chang ACY, Cohen SN. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.