Abstract
The identification of cardiac progenitor cells in mammals raises the possibility that the human heart contains a population of stem cells capable of generating cardiomyocytes and coronary vessels. The characterization of human cardiac stem cells (hCSCs) would have important clinical implications for the management of the failing heart. We have established the conditions for the isolation and expansion of c-kit-positive hCSCs from small samples of myocardium. Additionally, we have tested whether these cells have the ability to form functionally competent human myocardium after infarction in immunocompromised animals. Here, we report the identification in vitro of a class of human c-kit-positive cardiac cells that possess the fundamental properties of stem cells: they are self-renewing, clonogenic, and multipotent. hCSCs differentiate predominantly into cardiomyocytes and, to a lesser extent, into smooth muscle cells and endothelial cells. When locally injected in the infarcted myocardium of immunodeficient mice and immunosuppressed rats, hCSCs generate a chimeric heart, which contains human myocardium composed of myocytes, coronary resistance arterioles, and capillaries. The human myocardium is structurally and functionally integrated with the rodent myocardium and contributes to the performance of the infarcted heart. Differentiated human cardiac cells possess only one set of human sex chromosomes excluding cell fusion. The lack of cell fusion was confirmed by the Cre-lox strategy. Thus, hCSCs can be isolated and expanded in vitro for subsequent autologous regeneration of dead myocardium in patients affected by heart failure of ischemic and nonischemic origin.
Keywords: generation of human myocardium, progenitor cells, stem cell niches
The recent identification of different classes of cardiac progenitor cells has suggested that the heart is not a terminally differentiated, postmitotic organ but an organ regulated by a stem cell compartment (1). The possibility has also been raised that stem cells are present in the normal and pathological human heart (2, 3). Together, these results point to a shift in paradigm concerning the biology of the heart and put forward potential therapeutic strategies for the failing heart. However, the actual existence of a human cardiac stem cell (hCSC) remains to be demonstrated. By definition, stem cells have to be self-renewing, clonogenic, and multipotent in vitro and in vivo (4, 5), and no studies to date have shown that the human heart contains primitive cells with these properties. Cells with limited growth and differentiation ability may acquire only the myocyte, endothelial cell (EC) or smooth muscle cell (SMC) lineage in vitro, and may not be capable of forming functionally competent myocardium in vivo. hCSCs have to be able to replace dead tissue with contracting myocardium composed of cardiomyocytes and vascular structures, independently from cell fusion. Heterokaryons divide poorly and have, at best, a transient positive impact on the age of the fused cells (6). Here, we report that these issues have been resolved, and hCSCs may represent a form of therapy for the diseased heart.
Results
Cardiac Niches and hCSC Division.
The documentation of hCSCs requires the identification of interstitial structures with the characteristics of stem cell niches and the recognition of the mechanisms of stem cell division that regulate niche homeostasis and the self-renewing properties of the human heart in vivo (7). We have found that the human heart contains clusters of hCSCs that are intimately connected by gap junctions and adherens junctions to myocytes and fibroblasts (Fig. 1 A–C); myocytes and fibroblasts represent the supporting cells within the cardiac niches (7). Additionally, symmetric and asymmetric division of hCSCs was detected, respectively, by the uniform and nonuniform localization of the cell-fate determinants Numb and α-adaptin (7) at one or both poles of hCSCs in mitosis (Fig. 1 D and E). The commitment to the myocyte lineage of hCSCs was also found within the niches. The coexpression of the stem cell antigen c-kit and myocyte transcription factors and sarcomeric proteins [see supporting information (SI) Fig. 6] is consistent with a lineage relationship between hCSCs and myocyte formation. C-kitPOS cells expressing transcription factors for SMCs and ECs were also detected (data not shown). In the niches, hCSCs and committed cells were negative for hematopoietic markers and KDR (SI Table 1). These findings in the normal human heart, together with earlier observations in the diseased heart (3, 8), support the notion of a resident hCSC compartment that gives rise to the various cardiac cell progenies.
Fig. 1.
Cardiac niches and hCSC division. Sections of normal human myocardium. (A–C) Cluster of c-kitPOS cells (green). Arrows in A define the areas in B and C. Gap (connexin 43: Cx43, white; arrowheads) and adherens (N-cadherin: N-cadh, magenta; arrowheads) junctions are shown at higher magnification. Cx43 and N-cadh are present between c-kitPOS cells and myocytes (α-SA, red) and fibroblasts (procollagen, light blue); fibronectin, yellow. (D and E) Mitosis (phospho-H3, magenta; arrows) in c-kitPOS cells; α-adaptin (D, white) and Numb (E, yellow) show a uniform (D) and nonuniform (E) localization in the mitotic c-kitPOS cells.
Culture of hCSCs.
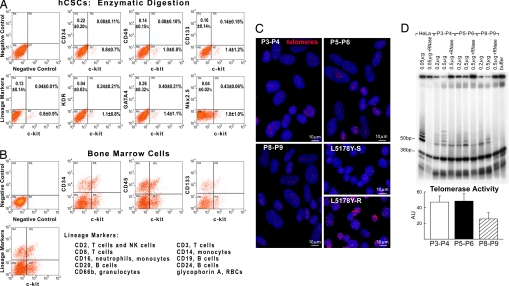
C-kitPOS cells, i.e., hCSCs, were prepared with two methodologies. The first consisted of the enzymatic dissociation of myocardial samples from which c-kitPOS cells were sorted with immunobeads and plated at low density (SI Fig. 7 A–C) to obtain multicellular clones from single founder cells. This procedure was dictated by the small size of the samples, ≈30 mg, which precluded FACS analysis at the outset. Successful isolation was obtained in 8 of 12 cases. The phenotype of the freshly isolated cells was characterized by FACS in 6 additional cases in which larger samples, ≈60 mg, were available. C-kitPOS cells comprised 1.1 ± 1.0% of the entire cell population. They were different from human bone marrow cells and were negative for markers of hematopoietic cells and KDR (Fig. 2 A and B; SI Table 2). Only small fractions of hCSCs expressed GATA4 and Nkx2.5, ≈0.5%.
Fig. 2.
Human CSCs. (A and B) Scatter plots of hCSCs (A) and human bone marrow cells (B). hCSCs do not express hematopoietic markers, KDR, GATA4, and Nkx2.5. (C) Nuclei (blue) of hCSCs were stained with a telomere probe (magenta). Lymphoma cells with short (7-kbp) and long (48-kbp) telomeres are shown for comparison. (D) Products of telomerase activity in hCSCs start at 50 bp and display a 6-bp periodicity. Samples treated with RNase and CHAPS buffer were used as negative controls, and HeLa cells were used as positive control. The band at 36 bp corresponds to an internal control for PCR efficiency. Optical density (arbitrary units, AU) is shown as mean ± SD.
With the second protocol, samples were cultured by the primary explant technique (SI Fig. 7 D and E). Successful cell outgrowth was obtained in 46 of 70 cases. A monolayer of ≈6,000 cells was present at the periphery of each tissue aggregate, ≈3 weeks after seeding. C-kitPOS cells accounted for 1.6 ± 1.4%. Adherent cells at passage P0 were analyzed by immunocytochemistry and FACS (SI Fig. 8; SI Tables 1 and 2). In enzymatically dissociated cells, lineage negative (Lin−) c-kitPOS cells were 41 ± 14%, and early committed cells (GATA4-positive) were 59 ± 14%. Corresponding values with the primary explant technique were 52 ± 12% and 48 ± 12% (SI Fig. 9A). In the presence of serum, hCSCs obtained with both protocols attached rapidly and continued to grow up to P8, undergoing ≈24 population doublings (PDs); the majority of experiments were concluded at P8. Cells maintained a stable phenotype and did not reach growth arrest. The percentage of c-kitPOS cells did not vary from P1 to P8, averaging 71 ± 8%. Undifferentiated cells constituted 63 ± 6%. Ki67POS cycling-cells averaged 48 ± 10%. p16INK4a, a cdk inhibitor, was present in 6 ± 4% of the cells (SI Fig. 9 B–D). Thus, hCSCs are distinct from bone marrow cells and can be isolated and expanded in vitro.
Telomere–Telomerase System.
To determine whether hCSCs reach senescence in culture, telomeric length was evaluated by Q-FISH (Fig. 2C). From P3–P4 (9–12 PDs) to P5–P6 (15–18 PDs) and P8–P9 (24–27 PDs), average telomere length in hCSCs decreased from 9.3 to 8.2 and 6.9 kbp, respectively (SI Fig. 10). From P3 to P9 there were ≈18 PDs with an average telomeric shortening of 130 bp per PD. This rate of telomere attrition is comparable with that of human bone marrow stem cells (9). Additionally, nearly 50% of the telomerase activity in hCSCs at P3–P4 was still present at P8–P9 (Fig. 2D).
Critical telomere length associated with growth arrest and cellular senescence of hCSCs and human hematopoietic SCs varies from 2.0 to 1.5 kbp (3, 9). The fraction of hCSCs with critical telomeric shortening increased from 1% at P3–P4 to 2% at P5–P6 and 5% at P8–P9. However, after ≈24–27 PDs at P8–P9, 69% hCSCs had telomere length ≥5.0 kbp (SI Fig. 10). It can be predicted that cells at P8–P9 can undergo 23 additional PDs (5–2 = 3kbp/0.13kbp = 23 PDs) before irreversible growth arrest (10). In theory, 50 PDs can result in the formation of 1 × 1015 hCSCs before replicative senescence is reached. Thus, hCSCs can be extensively grown in vitro in the absence of a major loss in their expansion potential.
hCSC Clones.
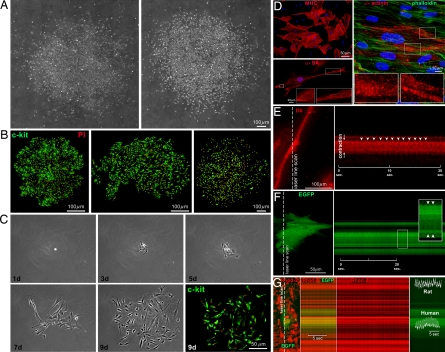
hCSCs obtained by enzymatic digestion and explant technique were plated at limiting dilution and in Terasaki plates, respectively. In the first case, 1,530 c-kitPOS cells were seeded, and after 3–4 weeks, 11 clones were generated. In the second case, cells were placed in individual wells, and 53 clones were formed from 6,700 seeded cells. Thus, hCSCs had ≈0.7–0.8% cloning efficiency (Fig. 3 A–C). Clones were expanded and characterized. Doubling time was 29 ± 10 h, and 90 ± 7% of cells after 5 days were BrdUPOS. Clonogenic hCSCs retained largely their primitive state and were negative for hematopoietic markers, KDR, and transcription factors and cytoplasmic proteins of cardiac cells (SI Fig. 11 A and B; SI Table 1).
Fig. 3.
In vitro properties of hCSCs. (A–C) Clones formed by hCSCs (c-kit, green) isolated by enzymatic digestion (A and C) or primary explant (B). The number of cells increased with time (C). (D) hCSCs generate myocytes positive for cardiac myosin heavy-chain (MHC), α-SA, and α-cardiac-actinin (α-actinin). Sarcomeres are apparent (Insets); phalloidin, green. (E) Myocyte shortening in cells derived from clonogenic hCSCs was recorded by two-photon microscopy and laser line-scan imaging (Left). The line scan is shown (Right), and arrowheads point to individual contractions. (F) Myocytes derived from EGFP-positive hCSCs, cocultured with neonatal myocytes. EGFP-positive human myocytes shorten (arrowheads) with electrical stimulation. (G) Calcium transients in EGFP-positive human myocytes and EGFP-negative rat myocytes (calcium indicator Rhod-2, red).
In differentiating medium, hCSCs gave rise to myocytes, SMCs, and ECs (Fig. 3D; SI Fig. 11 C and D). Developing myocytes had sarcomere striation (Fig. 3D) and, after electrical stimulation at 1 Hz, showed contractile activity (Fig. 3E). Moreover, hCSCs were infected with a lentivirus expressing EGFP and cocultured with neonatal rat myocytes. Two weeks later, cultures were stimulated, and 9% shortening of EGFP-positive human myocytes was detected (Fig. 3F). In the presence of the calcium indicator Rhod-2, calcium transient was identified in EGFP-positive human myocytes and EGFP-negative rat myocytes (Fig. 3G). Thus, hCSCs form multicellular clones and differentiate into contracting myocytes.
Human Myocardium.
Nonclonogenic hCSCs, collected from eight patients, were injected in the infarcted mouse or rat heart to form chimeric organs containing human myocytes and coronary vessels. Cell treatment led to areas of myocardial regeneration that were located within the infarct and were positive for α-sarcomeric actin (α-SA) and human AluDNA sequences (Fig. 4A). Human myocardium was found in 17 of 25 treated mice (68%), and 14 of 19 treated rats (74%). hCSCs were delivered with rhodamine-labeled microspheres for the recognition of the sites of injection and correct administration of cells (1). The absence of myocardial regeneration was due to technical failure to properly inject hCSCs in the rodent heart. Conversely, successful cell implantation was invariably associated with the presence of human myocardium.
Fig. 4.
hCSCs regenerate infarcted myocardium. (A) Mouse heart 21 days after infarction and injection of hCSCs. Human myocardium (arrowheads) is present within the infarct (MI). BZ, border zone. Areas in rectangles are shown at higher magnification below. Human myocytes are α-SA- (red) and Alu- (green) positive. Asterisks indicate spared myocytes. (B) Expression of human (h) genes by real-time RT-PCR in treated infarcted rats at 5–11 and 12–21 days. Clonogenic hCSCs were used for comparison of human transcripts. (C) Electrophoresis of real-time RT-PCR products (for sequences see SI Fig. 11J).
The human myocardium comprised 1.3 ± 0.9 mm3 in mice and 3.7 ± 2.9 mm3 in rats. Accumulation of new cells was also determined by BrdU labeling because BrdU was given to the animals throughout the experiment (SI Fig. 12 A and B). The human myocardium consisted of myocytes that occupied ≈84% of the tissue, whereas arterioles and capillaries accounted for ≈8%. The human origin of the myocardium was confirmed by the detection of human Alu and Mlc2v DNA by PCR in sections of regenerated infarcts (SI Fig. 12C). PCR products had the expected molecular weight, and the nucleotide sequences confirmed the specificity of the assay (SI Fig. 12 D–F).
Three control groups were used: (i) unsuccessfully treated-animals (eight mice, five rats); (ii) immunodeficient infarcted mice (n = 12) and immunosuppressed infarcted rats (n = 9) injected with PBS; and (iii) immunosuppressed infarcted rats injected with c-kit-negative cells obtained from the unfractionated cell population at P1 (n = 16). Infarct size was similar in all groups: 48 ± 9% in mice and 54 ± 11% in rats. Myocardial regeneration was absent in control hearts with the exception of 3 of the 16 hearts treated with c-kit-negative cells. In one case, a few α-SA and Alu-positive cells were found within the infarct, whereas, in the other two, a small band of human myocardium was identified near the border zone (SI Fig. 12 G and H).
For completeness, clonogenic hCSCs were injected in infarcted rats shortly after coronary ligation to determine their multipotentiality in vivo (n = 6) and establish whether multipotentiality persisted when cell implantation was performed 5 days after coronary occlusion under the condition of a fully developed ischemic injury (n = 10). In both cases, clonogenic hCSCs regenerated the infarcted myocardium (SI Fig. 12I) by forming human myocytes and coronary vessels (see below).
Real-time RT-PCR was used to demonstrate human transcripts for myocyte (MLC2v, connexin 43), SMC (smooth-muscle myosin heavy-chain 11 = Mhc 11) and EC (vWF) genes, and the housekeeping gene GAPDH in infarcted rat hearts treated with clonogenic hCSCs. Because there is no baseline in the rat myocardium for the analysis of human genes, clonogenic hCSCs were used for comparison (n = 4). With respect to clonogenic hCSCs, there was a substantial up-regulation of human myocardial transcripts for parenchymal and vascular cells in the infarcted heart (Fig. 4B). The expression of human MLC2v, connexin 43, Mhc 11, and vWF increased from 5–11 days (n = 8) to 12–21 days (n = 15) after infarction and cell implantation. RT-PCR products had the expected molecular weight (Fig. 4C), and the nucleotide sequences confirmed the specificity of the assay (SI Fig. 12J). Thus, hCSCs generate human myocardium.
Regenerated Myocardium.
After the identification of AluDNA, cardiac myosin heavy-chain (MHC), troponin I, and α-SA were detected in human myocytes. Moreover, GATA4, MEF2C, connexin 43, and N-cadherin were identified (SI Figs. 13 and 14). Human myocytes varied in size from 100 to 2,900 μm3 (SI Fig. 14). Human coronary arterioles and capillaries were also found (SI Figs. 13 and 14). The number of human arterioles and capillaries was comparable in rats and mice; there was one capillary/eight myocytes (SI Fig. 14), and the diffusion distance for oxygen averaged 18 μm. These parameters are similar to those found in the late fetal–neonatal human heart. Thus, hCSCs differentiate into human myocytes and coronary vessels, leading to the formation of a chimeric heart.
Cell Fusion.
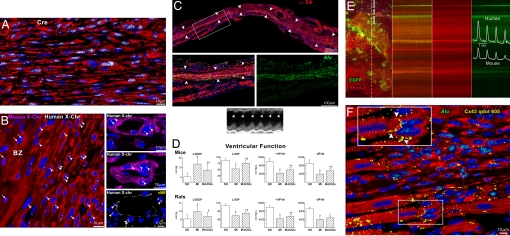
Two protocols were used to test whether the generation of human myocardium involved fusion events between hCSCs and rodent cells. hCSCs were infected with a lentivirus carrying Cre-recombinase (infection efficiency = 90%) and injected in the infarcted heart of mice expressing loxP-flanked EGFP (n = 6). If fusion were to occur, EGFP transcription would be activated in the recipient cells by Cre-mediated excision of the stop codon in the EGFP promoter (1). At 10 days after infarction and cell implantation, newly formed human myocardial cells showed a nuclear localization of Cre protein (Fig. 5A; SI Fig. 15). However, human myocytes and vessels were negative for EGFP, indicating that the formation of heterokaryons was not involved in cardiac repair.
Fig. 5.
Integration of human myocardium. (A) Human myocytes are Cre-recombinase-positive (white) but EGFP negative. (B) Human myocytes and vessels show, at most, two human X-chromosomes (X-Chr, white dots; arrowheads). Mouse X-Chr (magenta dots; arrows) are present in myocytes of the border zone (BZ). (C) Transmural infarct in a treated rat; human myocardium (arrowheads) is present within the infarct. The area in the rectangle is shown at higher magnification (Bottom); human myocytes are α-SA- (red) and Alu- (green) positive. Echocardiogram shows contraction in the infarcted wall (arrowheads). (D) Ventricular function. Results are mean ± SD. * and †, Difference, P < 0.05, versus SO (sham-operated) and MI, respectively. (E) Calcium transient in EGFP-positive human myocytes and EGFP-negative mouse myocytes recorded by two-photon microscopy and laser line-scan imaging (calcium indicator Rhod-2, red). (F) Myocardial regeneration at 3 weeks. Connexin 43 (Cx43, yellow) is present between human-myocytes (α-SA, red; Alu, green) and spared rat myocytes (α-SA, red; Alu-negative). See Inset for higher magnification.
The second protocol consisted of the evaluation of the number of sex chromosomes by Q-FISH in human myocytes and coronary vessels. Because female human cells were injected in female mice and rats, human, rat and mouse X-chromosomes were measured. We never found a colocalization of a human X-chromosome with a mouse or rat X-chromosome in regenerated myocytes and vessels (Fig. 5B). Human myocytes, SMCs, and ECs carried, at most, two human X-chromosomes. Thus, hCSCs form human myocardium independently from cell fusion.
Functional Competence of Human Myocardium.
Echocardiograms were examined retrospectively after the histological documentation of transmural infarcts and the presence of human myocardium (Fig. 5C; SI Fig. 16 A–C). Tissue regeneration restored partly contractile function in the infarct, resulting in an increase of ejection fraction (SI Fig. 16D), attenuation of chamber dilation (SI Fig. 16E), and improvement of ventricular function (Fig. 5D).
The interaction between human and rodent myocardium was determined by an ex vivo preparation and two-photon microscopy. EGFP-positive hCSCs were injected in infarcted mice, and the heart was studied 2 weeks later (n = 6). After the blockade of contraction and spontaneous activity, the heart was perfused with Rhod-2 and stimulated at 1 Hz. Calcium transient was recorded in EGFP-positive human myocytes and EGFP-negative mouse myocytes. The synchronicity in calcium tracings between these myocyte populations documented their functional integration (Fig. 5E). hCSC-derived myocytes acquired the properties of the recipient rodent myocardium, indicating that primitive cells of human origin possess a high level of plasticity. Additionally, connexin 43 was found between human and rodent myocytes (Fig. 5F) demonstrating their structural coupling. Thus, both myocardial components of the chimeric heart participate in the performance of the infarcted heart.
Discussion
The current work demonstrates that the human heart possesses a pool of clonogenic hCSCs that can acquire the myocyte, SMC, and EC lineages in vitro and in vivo. The ability of hCSCs to create cardiomyocytes and coronary vessels in vivo provides strong evidence in favor of the role that hCSCs have in cardiac homeostasis and myocardial regeneration. Besides their therapeutic implications, these observations challenge the view of the heart as a postmitotic organ (11) and form the basis of a paradigm in which multipotent hCSCs modulate the physiological turnover of the heart. Understanding the mechanisms of cardiac homeostasis would offer the opportunity to potentiate this process and promote cardiac repair after injury.
Human cells with the ability to differentiate into cardiomyocytes have been obtained from myocardial biopsies and were claimed to possess the properties of stem cells (2). These cells express the typical markers of human circulating endothelial-progenitor cells (EPCs): CD34, CD31, and KDR, together with c-kit (12). The expression of CD34, CD31, and KDR does not compromise the ability of these circulating cells to acquire the myocyte lineage in vitro (13) and in vivo (14). The presence of these epitopes, however, suggests that these cells originate from the bone marrow and only subsequently accumulate within the heart. These early findings failed to provide evidence for the clonogenicity of these cells in vitro and their multilineage differentiation in vivo, which are critical for the recognition of a tissue-specific adult stem cell (1). The inability of these cells to generate a functional human myocardium in vivo is consistent with the role of EPCs in cardiac repair; they acquire, at low efficiency, the myocyte lineage and exert a paracrine effect on the infarcted heart (13).
Conversely, as demonstrated here, hCSCs are positive for the stem cell antigen c-kit but are negative for the hematopoietic and endothelial antigens CD45, CD34, CD31, and KDR; CD45 and KDR are typically expressed in a subset of bone marrow c-kitPOS cells that have the ability to migrate to the heart after injury (12). Stem cell niches have been identified here in the normal human myocardium, and hCSCs divide symmetrically and asymmetrically and give rise to differentiating and lineage-negative cells. This provides evidence in favor of a linear relationship between hCSCs and myocyte formation. Additionally, these observations do not support the notion of dedifferentiation of mature myocytes with the acquisition of a stem cell phenotype. Importantly, clonogenic hCSCs have the inherent potential to form contracting myocardium integrated structurally and functionally with the recipient heart. Although CSCs with similar characteristics were shown in animal models (4, 5), the applicability of this information to humans was seriously questioned and considered a major limitation for the clinical implementation of CSCs (15).
In the current study, three possibilities were considered in the formation of human myocardium within the infarcted mouse and rat heart (1): (i) Growth and differentiation of hCSCs; (ii) Fusion of hCSCs with the surviving mouse or rat cardiac cells, followed by proliferation of the heterokaryons and generation of myocytes and coronary vessels; and (iii) A combination of these two processes. The evaluation of human, mouse, and rat sex chromosomes together with the Cre-lox strategy has indicated that the generation of human myocardium involved only the commitment of hCSCs to cardiomyocytes, SMCs, and ECs. The unlikely involvement of cell fusion was supported by the size (100–2,900 μm3) of human myocytes. If fusion were to be implicated, the newly formed human myocytes should have had a volume of at least 25,000 μm3 or larger, that is, the volume of adult mouse and rat cardiomyocytes. It is improbable that fusion of a primitive cell with a terminally differentiated myocyte can induce division of a highly specialized and rapidly contracting cell permanently withdrawn from the cell cycle (1, 6).
The identification of a resident hCSC pool in the human heart is apparently at variance with the small foci of myocardial regeneration present after acute and chronic infarcts or pressure overload in patients (3, 16). The limitation that resident hCSCs have in reconstituting myocardium after infarction has been interpreted as the unequivocal documentation of the inability of the adult heart to create cardiomyocytes (11). The inevitable evolution of ischemic injury is myocardial scarring with loss of mass and contractile function. A possible explanation of this apparent paradox has been obtained in animal models of the human disease (5). Stem cells are present throughout the infarcted myocardium but, despite the postulated resistance of these cells to death stimuli, they follow the same pathway of cardiomyocytes and die by apoptosis. The fate of hCSCs is comparable with that of the other cells, and myocyte formation is restricted to the viable portion of the human heart (3).
It might come as a surprise, but a similar phenomenon occurs in solid and nonsolid organs, including the skin, liver, intestine, and kidney. In all cases, occlusion of a supplying artery leads to scar formation mimicking cardiac pathology (17–20). In the presence of polyarteritis nodosa and vasculitis, microinfarcts develop in the intestine and skin, and resident SCs do not repair the damaged organs (21). In nonsolid organs, infarcts of the bone marrow are seen with sickle cell anemia (21). Thus, the SC compartment appears to be properly equipped to modulate growth during postnatal development and regulate homeostasis in adulthood. However, SCs do not respond effectively to ischemic injury or, late in life, to aging and senescence of the organ and organism (22, 23).
Current knowledge supports the notion that primitive cells are present in the heart at the very beginning of embryonic life and regulate heart morphogenesis and postnatal development (24). By introducing the EGFP gene in the mouse embryo, at the stage of the morula–blastocyst transition, the patterns of myocardial histogenesis have been defined and the presence of a common progenitor of cardiomyocytes in prenatal and postnatal life suggested (24). The documentation of myocardial specification of embryonic stem cells (25, 26), in particular c-kitPOS Nkx2.5POS cells (26), supports the view that a pool of resident c-kitPOS progenitors is implicated in cardiac morphogenesis. These findings are consistent with the existence of a pool of primitive cells in the adult human heart.
Materials and Methods
hCSCs have been isolated expanded and characterized in vitro and in vivo after implantation in the infarcted rodent heart. Protocols are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
The lentivirus expressing Cre-recombinase was kindly provided by Drs. Chang and Terada (University of Florida, Gainesville, FL) and the lymphoma cells by Dr. Blasco (Spanish National Cancer Centre, Madrid, Spain). This work was supported by National Institutes of Health grants.
Abbreviations
- EC
endothelial cell
- hCSC
human cardiac stem cell
- PD
population doubling
- SMC
smooth muscle cell.
Footnotes
Conflict of interest statement: P.A. has applied for a patent.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706760104/DC1.
References
- 1.Leri A, Kajstura J, Anversa P. Physiol Rev. 2005;85:1373–1416. doi: 10.1152/physrev.00013.2005. [DOI] [PubMed] [Google Scholar]
- 2.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, Salio M, Battaglia M, Latronico MVG, Coletta M, et al. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 3.Urbanek K, Torella D, Sheikh F, De Angelis A, Nurzynska D, Silvestri F, Beltrami CA, Bussani R, Beltrami AP, Quaini F, et al. Proc Natl Acad Sci USA. 2005;102:8692–8697. doi: 10.1073/pnas.0500169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, et al. Cell. 2003;114:763–766. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 5.Linke A, Muller P, Nurzynska D, Casarsa C, Torella D, Nascimbene A, Castaldo C, Cascapera S, Bohm M, Quaini F, et al. Proc Natl Acad Sci USA. 2005;102:8966–8971. doi: 10.1073/pnas.0502678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weimann JM, Johansson CB, Trejo A, Blau HM. Nat Cell Biol. 2003;5:959–966. doi: 10.1038/ncb1053. [DOI] [PubMed] [Google Scholar]
- 7.Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T, Bearzi C, Boni A, Bolli R, Kajstura J, et al. Proc Natl Acad Sci USA. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quaini F, Urbanek K, Beltrami AP, Finato N, Beltrami CA, Nadal-Ginard B, Kajstura J, Leri A, Anversa P. N Engl J Med. 2002;346:5–15. doi: 10.1056/NEJMoa012081. [DOI] [PubMed] [Google Scholar]
- 9.Van Ziffle JAG, Baerlocher GM, Lansdorp PM. Stem Cells. 2003;21:654–660. doi: 10.1634/stemcells.21-6-654. [DOI] [PubMed] [Google Scholar]
- 10.Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Nat Biotech. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 11.Chien KR. Nature. 2004;418:607–608. doi: 10.1038/nature02500. [DOI] [PubMed] [Google Scholar]
- 12.Fazel S, Cimini M, Chen L, Li S, Angoulvant D, Fedak P, Verma S, Weisel RD, Keating A, Li RK. J Clin Invest. 2006;116:1865–1877. doi: 10.1172/JCI27019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badorff C, Brandes RP, Popp R, Rupp S, Urbich C, Aicher A, Fleming I, Busse R, Zeiher AM, Dimmeler S. Circulation. 2003;107:1024–1032. doi: 10.1161/01.cir.0000051460.85800.bb. [DOI] [PubMed] [Google Scholar]
- 14.Murasawa S, Kawamoto A, Horii M, Nakamori S, Asahara T. Arterioscler Thromb Vasc Biol. 2005;25:1388–1394. doi: 10.1161/01.ATV.0000168409.69960.e9. [DOI] [PubMed] [Google Scholar]
- 15.Murry CE, Field LJ, Menasche P. Circulation. 2005;112:3174–3183. doi: 10.1161/CIRCULATIONAHA.105.546218. [DOI] [PubMed] [Google Scholar]
- 16.Urbanek K, Quaini F, Tasca G, Torella D, Castaldo C, Nadal-Ginard B, Leri A, Kajstura J, Quaini E, Anversa P. Proc Natl Acad Sci USA. 2003;100:10440–10445. doi: 10.1073/pnas.1832855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez LR, Schocket AL, Stanford RE, Claman HN, Kohler PF. J Rheumatol. 1980;7:677–684. [PubMed] [Google Scholar]
- 18.Saegusa M, Takano Y, Okudaira M. Liver. 1993;13:239–245. doi: 10.1111/j.1600-0676.1993.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe K, Abe H, Mishima T, Ogura G, Suzuki T. Pathol Int. 2003;53:569–573. doi: 10.1046/j.1440-1827.2003.01515.x. [DOI] [PubMed] [Google Scholar]
- 20.Leong FT, Freeman LJ. J R Soc Med. 2005;98:121–122. doi: 10.1258/jrsm.98.3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang NC, Johnson C, Eslami-Farsani M, Haywood LJ. Am J Hematol. 2005;79:61–67. doi: 10.1002/ajh.20348. [DOI] [PubMed] [Google Scholar]
- 22.Chimenti C, Kajstura J, Torella D, Urbanek K, Heleniak H, Colussi C, Di Meglio F, Nadal-Ginard B, Frustaci A, Leri A, et al. Circ Res. 2003;93:604–613. doi: 10.1161/01.RES.0000093985.76901.AF. [DOI] [PubMed] [Google Scholar]
- 23.Rossi DJ, Bryder D, Zahn JM, Alhenius H, Sonu R, Wagers AJ, Weissman IL. Proc Natl Acad Sci USA. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberhard D, Jockusch H. Dev Biol. 2005;278:336–346. doi: 10.1016/j.ydbio.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Kattman SJ, Huber TL, Keller GM. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, Fujiwara Y, Cibulsky S, Clapham D, Lien C, Schultheiss T, Orkin S. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.