Abstract
We report a suite of key microfluidic devices for complex high-throughput whole-animal genetic and drug screens. We demonstrate a high-speed microfluidic sorter that can isolate and immobilize Caenorhabditis elegans in a well defined geometry for screening phenotypic features at subcellular resolution in physiologically active animals. We show an integrated chip containing individually addressable screening-chamber devices for incubation and exposure of individual animals to biochemical compounds and high-resolution time-lapse imaging of many animals on a single chip without the need for anesthesia. We describe a design for delivery of compound libraries in standard multiwell plates to microfluidic devices and also for rapid dispensing of screened animals into multiwell plates. When used in various combinations, these devices will facilitate a variety of high-throughput assays using whole animals, including mutagenesis and RNAi and drug screens at subcellular resolution, as well as high-throughput high-precision manipulations such as femtosecond laser microsurgery for large-scale in vivo neural degeneration and regeneration studies.
Keywords: Caenorhabditis elegans, femtosecond laser microsurgery, immobilization and time-lapse imaging, mutagenesis, RNAi and drug screening
Existing large vertebrate animal models currently cannot be used in high-throughput assays for rapid identification of new genes and drug targets because of the size and complexity of the instrumentation with which these models are studied. In recent years, the advantages of using small invertebrate animals as model systems for human disease have become increasingly apparent and have resulted in two Nobel Prizes in physiology and medicine during the last five years for studies conducted on the nematode Caenorhabditis elegans. The availability of a wide array of species-specific genetic techniques, along with the transparency of the worm and its ability to grow in minute volumes make C. elegans an extremely powerful model organism.
However, since the first studies on C. elegans in the early 1960s, little has changed in how scientists manipulate this tiny organism by manually picking, sorting, and transferring individual animals. As a result, large-scale assays such as mutagenesis and RNAi screens (1–3) can take months or even years to complete manually. Currently, high-throughput C. elegans assays are performed by adapting techniques developed for screening cell lines, such as flow-through sorters and microplate readers (4–6). Because of the significant limitations of these methods, high-throughput small-animal studies either have to be dramatically simplified before they can be automated or cannot be conducted at all.
Here, we report key components of an integrated, whole-animal, high-throughput sorting and large-scale screening platform for drug and genetic assays with subcellular resolution using microfluidic devices. Although microfluidics have previously been used to perform novel assays on C. elegans, so far research has been limited to specific applications such as generation of oxygen gradients (7), worm culturing/monitoring during spaceflight (8), optofluidic imaging (9), and maze exploration (10). We have designed microfluidic devices that can be combined in various configurations to allow a multitude of complex high-throughput assays such as mutagenesis, drug and RNAi screens: (i) a small-animal sorter for sorting live animals according to subcellular features; (ii) an array of microfluidic chambers for simultaneous incubation, immobilization, subcellular-resolution imaging and independent screening of many animals on a single chip; and (iii) a microfluidic interface to large-scale multiwell-format libraries that also functions as a multiplexed animal dispenser.
The microfluidic devices that we fabricated consist of flow and control layers made from flexible polymers (11). The flow layers contain microchannels for manipulating C. elegans, immobilizing them for imaging, and delivering media and reagents. The flow layers also contain microchambers for incubating the animals. The control layers consist of microchannels that when pressurized, flex a membrane into the flow channels, blocking or redirecting the flow (12). Animals in the flow lines can be imaged through a transparent glass substrate using high-resolution microscopy.
On-Chip High-Throughput Sorting.
Sorters enable rapid selection of organisms with phenotypes of interest for a variety of assays, including genetic and drug screens, and also for reducing phenotypic variability in large-scale assays. Existing small-animal sorters such as the BIOSORT/COPAS machine, use a flow-through technique similar to the fluorescence-activated cell sorter (FACS) technology. These systems can capture and analyze only one-dimensional intensity profiles of the animals being sorted, and as a result, three-dimensional cellular and subcellular features cannot be resolved (13). To address this problem and to achieve on-chip integration we have developed the animal sorter shown in Fig. 1. Animals enter the chip through the inlet channel and can be continuously recirculated. A single worm is captured in an immobilization chamber via suction by a microchannel held at a low pressure. The use of a single suction channel eliminates the problem of simultaneously capturing multiple animals. While the captured animal is held in the immobilization chamber, all of the other animals in the chamber are removed by flushing with media from a side channel. This step ensures that only a single animal is isolated even when the concentration of worms is high. The animals that are flushed in our present design could be recirculated for screening if needed. Next, valves are closed to isolate the chamber containing the single worm from the rest of the chip. The captured worm is then released from the single suction channel and recaptured by an array of suction channels to restrain it in a straight position. At this stage, the worm can be imaged through the transparent glass substrate by using high-resolution optics for phenotype analysis (Fig. 2) [see supporting information (SI) movie 1]. The chip is designed to allow both morphological details and fluorescence markers to be detected with white-light and epifluorescence imaging with subcellular resolution (Fig. 2). Three-dimensional cross-sectioning by two-photon microscopy could also be used at the expense of sorting speed. After image acquisition and processing, the captured worm can be released and directed to the appropriate collection channels according to its phenotype.
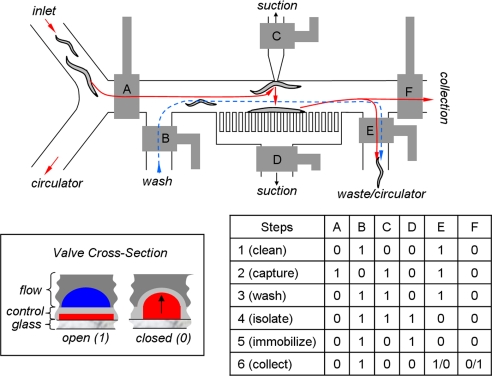
Fig. 1.
Microfluidic worm-sorter layout and operation. The sorter consists of control channels and valves (gray) that direct the flow of worms in the flow channels in different directions. The valves are labeled with the letters A–F in the layout, and the actuation order of valves is listed in the table. A value of 1 represents an open valve, and a value of 0 represents a closed valve, as illustrated in the lower-left box. The steps taken to sort each worm are as follows: step 1 (clean), the immobilization chamber is cleaned; step 2 (capture), a worm is captured in the chamber by suction via the top channel while the lower suction channels are inactive; step 3 (wash), the chamber is washed to flush any other worms in the chamber (blue line) toward the waste or the circulator; step 4 (isolate), the chamber is isolated from all of the channels; step 5 (immobilize), the worm is released from the top suction channel and is restrained by the lower suction channels; step 6 (collect), the image acquisition and processing are performed, and the worm is either collected or directed to the waste, depending on its phenotype.
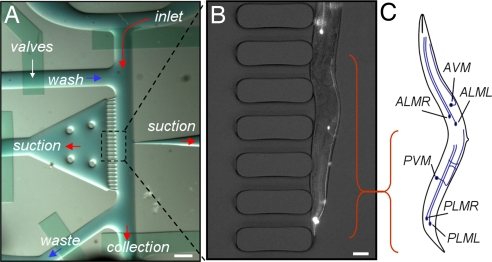
Fig. 2.
Immobilization and subcellular imaging using worm sorter. (A) Image of the on-chip sorter described in Fig. 1. (Scale bar: 500 μm.) (B) A single worm is shown trapped by multiple suction channels. A combined white-light and fluorescence image is taken by a cooled CCD camera with 6.5-μm pixels and a 100-ms exposure time through a ×10 magnification, 0.45 N.A. objective lens with (Nikon). mec-4::GFP-expressing touch neurons and their processes are clearly visible. (Scale bar: 10 μm.) (C) The mechanosensory neurons PLML/R and ALML/R (L, left; R, right) are shown. AVM and PVM extend processes along the anterior and posterior half of the worm and contribute to mechanosensation in these regions. The cell bodies are shown as black dots. PVM, posterior ventral mechanosensory; ALM, anterior lateral mechanosensory; AVM, anterior ventral mechanosensory.
The microfluidic chips have flow and control layers and are permanently bonded onto glass substrates to allow optical access. Flow layers are made by casting a room-temperature-vulcanizing dimethylsiloxane polymer (RTV615; GE Silicones, Wilton, CT) by using a mold consisting of a patterned layer of positive photoresist (SIPR-7123; Shin-Etsu, Tokyo, Japan) on a silicon wafer. Flow layer channels are 250–500 μm wide and 80–110 μm high. The channels are rounded by reflowing the developed photoresist at 150°C. In the current design, the flow layer is made from a mold with a single photoresist layer that defines suction channels that are 40 μm high and 50 μm wide after reflow, which allows capturing of adult worms. To capture juvenile worms, a two-layer photoresist mold could be used to make smaller suction channels. Control layers are made by casting from a mold consisting of a patterned layer of negative photoresist (SU-8 2075; MicroChem, Newton, MA) on a silicon wafer. Control channels are 70–80 μm high, and the membrane that separates the two layers is 10–20 μm thick. Polydimethylsiloxane chips cost significantly less than current flow-through animal-screening machines and can be easily incorporated into a variety of microscopy systems.
The speed of the sorter depends on the actuation speed of the valves, the concentration of animals at the input, the flow speed of the worms, and the image acquisition and processing times. The technique of immobilizing worms by lowering pressure in a microchannel is fast because the actuation speed of the valves is <30 ms. Because of the continuous recirculation at the input, animals can be flowed at high concentration without clogging the chip. The speed of image acquisition and recognition of subcellular features is fundamentally limited by the fluorescence signal-to-noise ratio and the complexity of the features being recognized. The entire worm can be imaged in a single frame by using a low magnification, high-N.A. objective lens. Cellular and subcellular features (touch-neuron axons, etc.) can be detected by wide-field epifluorescence, where the exposure times are limited by the brightness of the fluorescent markers. Using a cooled CCD camera (Roper Scientific, Trenton, NJ), we were able to perform image acquisition at speeds exceeding one frame every 100 ms when imaging neurons labeled with green fluorescent protein (GFP). As a result of these features, this design can allow sorting of worms at high speeds.
Large-Scale Time-Lapse Assays with Subcellular Resolution.
Time-lapse imaging is important for a variety of assays, including drug and genetic screens. Currently, high-throughput time-lapse studies on small animals are done in multiwell plates by automated fluorescence microplate readers (4). Because the animals swim inside the wells, only average fluorescence is obtained from each well, and cellular and subcellular details cannot be imaged. Although anesthesia can be used to immobilize the animals, they cannot be kept under anesthesia for more than a few hours, and they cannot be anesthetized frequently. Furthermore, the effect of anesthesia on many biological processes remains uncharacterized. Another limitation of current multiwell plates is the loss of animals that occurs during media exchange. To address these problems, we designed the microfluidic-chamber device shown in Fig. 3A for worm incubation and for continuous imaging at subcellular resolution. Sorted worms can be delivered to the chambers by opening valves via multiplexed control lines (14). The fabrication of the channels and valves is done as described earlier. To ensure that the chamber structure is not affected by the reflow process, two types of photoresist are used to make the mold for the flow layer. The chambers are first fabricated by using a 100-μm-thick layer of SU8-2075 (which does not reflow), and then the valve-controlled flow channnels are fabricated on the same water by using a 100-μm-thick layer of SIPR-7123. The pressure in the control lines is switched on and off with external electronically controlled valves (Numatics, Highland, MI). Because the number of control lines required to independently address n incubation chambers scales only with log(n) (14), microchamber chips based on this design can be readily scaled for large-scale screening applications. Because of the millimeter scale of the microchambers, hundreds of microchambers could be integrated on a single chip. Each incubation chamber contains posts arranged in an arc. To image animals, a flow is used to push the animals toward the posts (Fig. 3 B and C). This flow restrains the animals for subcellular imaging. The circular arrangement of the posts reduces the size of the chambers and also positions the animals in a well defined geometry to reduce the complexity and processing time of image recognition algorithms. The medium in the chambers can be exchanged through the microfluidic channels for complex screening strategies. Thus, precisely timed exposures to biochemicals (e.g., drugs/RNAi) can be performed, which is useful both for identifying mechanisms that rely on the action of more than one compound and for combinatorial assays involving multiple drug targets. The use of microfluidic technology also reduces the cost of whole-animal assays by reducing the required volumes of compounds.
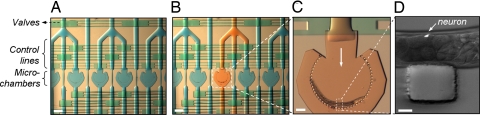
Fig. 3.
Microchamber chip for large-scale screening. (A) The chip consists of chambers connected to flow lines in which the flow path is controlled via multiplexed control lines and valves. (Scale bar: 500 μm.) (B) Each chamber can be addressed independently and loaded with compounds. The flow lines can be flushed with a wash buffer through a dedicated line to prevent cross-contamination. (Scale bar: 500 μm.) (C) The same flow lines can also be used to deliver worms. A special microchamber geometry that consists of circularly arranged microposts is used to immobilize the animals quickly in a well defined geometry by applying a flow without using anesthetics. (Scale bar: 100 μm.) (D) High-resolution images can be taken through the glass substrate of the chip. The GFP-labeled fluorescent touch-neuron image was taken with a white-light background to show a micropost. (Scale bar: 25 μm.) (A–C) Images were taken using a stereomicroscope after loading fluidic lines with color dyes. (D) Image was taken in another setup using an inverted fluorescence microscope.
Microfluidic Multiwell-Plate Interface Chip for Compound-Library Delivery and Multiplexed Animal Dispensing.
Interfacing microfluidics to existing large-scale RNAi and drug libraries in standard multiwell plates represents a significant challenge. It is impractical to deliver compounds to thousands of microchambers on a single chip through thousands of external fluidic connectors. To address this problem, we designed the microfluidic interface chip shown in Fig. 4. The device consists of an array of aspiration tips that can be lowered into the wells of microwell plates. The chip is designed to allow minute amounts of library compounds to be collected from the wells by suction, routed through multiplexed flow lines one at a time, and delivered to the single output of the device. The output of the interface chip can then be connected to our microfluidic-chamber device for sequential delivery of compounds to each microchamber. Combining this multiwell-plate interface chip with existing robotic multiwell-plate handlers will allow large libraries to be delivered to microfluidic chips. The same device can also be used to dispense worms into multiwell plates, simply by running it in reverse.
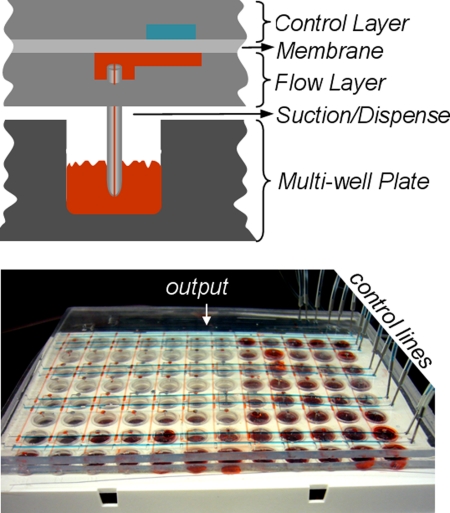
Fig. 4.
Design for delivery of compounds from standard multiwell plates to microfluidic devices. A microfluidic chip loads compounds from multiwell plates to flow channels by aspiration. The flow lines are multiplexed (14) to direct one compound at a time to a single serial output. The direction of flow in the channels is controlled by microfluidic valves as described in Fig. 1. The flow lines are flushed with a wash buffer after loading each compound to prevent cross-contamination. The single serial output of this device can easily be connected to the microchamber screening chip (Fig. 3) for compound delivery. Each microchamber chip is also multiplexed (14) to sort and deliver compounds to individual chambers (Fig. 3).
Discussion
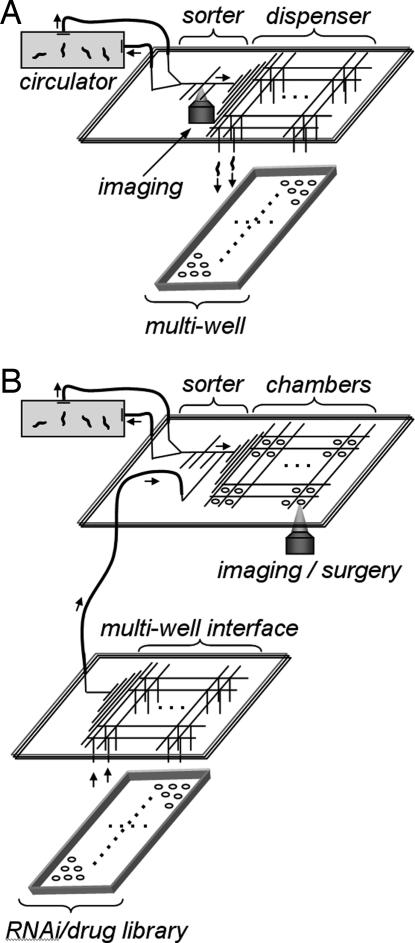
Because our sorter and microchambers are designed to immobilize and release animals repeatedly in <100 ms, the on-chip screening technology that we introduced here will allow high-throughput whole-animal assays at subcellular resolution and with time-lapse imaging in physiologically active animals. It will be possible to automate a variety of assays by combining our devices in different configurations. Mutagenesis screens could be performed by using our microfluidic sorter in combination with our microfluidic dispenser to dispense sorted animals at high speeds into the wells of multiwell plates (Fig. 5A). Large-scale RNAi and drug screens with time-lapse imaging could be performed by combining our sorter, integrated microchambers, and multiwell plate interface chips as shown in Fig. 5B. Although C. elegans is self-fertilizing and has perhaps the lowest phenotypic variability among multicellular model organisms (4), variations among assayed animals are still present, reducing the robustness of current large-scale screens. Sorting technology can be used to select animals with similar phenotypes (such as fluorescent marker expression levels) before large-scale assays to significantly reduce initial phenotypic variations (Fig. 5B) (4, 15). Our microchamber technology could be used with feature-extraction algorithms to screen thousands of animals on a single chip. An interface to multiwell plates can be used to deliver large compound libraries to our microchambers.
Fig. 5.
Possible combinations of our microfluidic technologies for large-scale high-throughput assays. (A) High-speed phenotype screens (e.g., after genetic mutagenesis) can be performed at cellular or subcellular resolution by cascading the microfluidic sorter with the multiwell dispenser. (B) Large-scale RNAi/drug screens can be performed by delivering standard multiwell plate libraries to the microfluidic screening chambers via the multiwell interface chips.
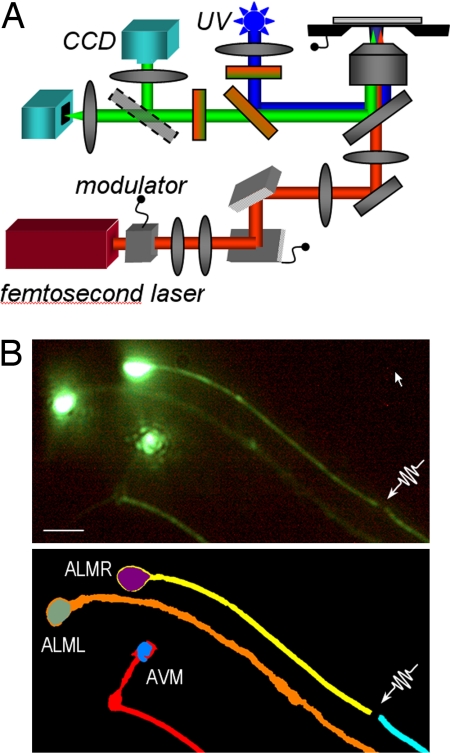
Recently, Yanik et al. (16, 17) demonstrated femtosecond laser microsurgery in C. elegans as a precise and reproducible injury model to study neural degeneration and regeneration in vivo. The immobilization technique described above will allow us to hold the worms still during such precision microsurgical operations (Fig. 5B) and perform time-lapse imaging at subcellular resolution after surgery. Fig. 6 shows a feature-extracted image of a worm acquired at cellular resolution after performing femtosecond laser microsurgery. Time series of such images could be used to measure growth-cone movement rates, the direction of outgrowth relative to the original trajectory, and the degree of branching for high-throughput, whole-animal studies of neural degeneration and regeneration after injury. Because the immobilized animals are physiologically active and not anesthetized, their neural activity patterns can potentially be imaged at cellular resolution by using genetically encoded optical probes.
Fig. 6.
Femtosecond laser microsurgery of axons. (A) Optical setup for delivery of femtosecond pulses to a specimen through a high-N.A. objective lens. (B) (Upper) Fluorescence images of mec4::GFP-labeled touch neurons (ALMR, ALML, AVM). L and R, left and right. Femtosecond laser microsurgery is performed on the target process with 100-fs, 3-nJ pulses at a repetition rate of 80 MHz. (Lower) Cell bodies and neural processes identified after edge detection and feature extraction. (Scale bar: 5 μm.) Color coding shows individual cell bodies, neural processes, and the laser cut. Feature extraction was performed by thresholding first to identify cellular features and then, combined with a Canny edge detection algorithm, to identify the outline of neural processes. AVM, anterior ventral mechanosensory; ALM, anterior lateral mechanosensory; L, left; R, right.
The key integrated high-throughput technologies that we report here has the potential not only to significantly accelerate current whole-animal genetic and drug screens but also to enable completely new types of assays at subcellular resolution.
Supplementary Material
Acknowledgments
We thank the members of the Steve Quake laboratory at Stanford University, in particular J. Melin, F. Balagadde, and R. Gomez-Sjoberg, for advice on fabrication; and the members of the Robert Horvitz laboratory at Massachusetts Institute of Technology, in particular R. O'Hagan and N. Ringstad, for help on C. elegans. F.Z. and R.G.-R. were supported by fellowships from the Canadian National Science and Engineering Research Council and the Paul and Daisy Soros Foundation, respectively.
Footnotes
Conflict of interest statement: U.S. and international patents filed on small-animal sorting and screening technologies.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706513104/DC1.
References
- 1.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 2.Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sieburth D, Ch'ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, et al. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 4.Kaletta T, Butler L, Bogaert T. Model Organisms in Drug Discovery. West Sussex, UK: Wiley; 2003. [Google Scholar]
- 5.Kaletta T, Hengartner MO. Nat Rev Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 6.Segalat L. ACS Chem Biol. 2007;2:231–236. doi: 10.1021/cb700009m. [DOI] [PubMed] [Google Scholar]
- 7.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 8.Lange D, Storment C, Conley C, Kovacs G. Sensor Actuator B Chem. 2005;107:904–914. [Google Scholar]
- 9.Heng X, Erickson D, Baugh LR, Yaqoob Z, Sternberg PW, Psaltis D, Yang C. Lab Chip. 2006;6:1274–1276. doi: 10.1039/b604676b. [DOI] [PubMed] [Google Scholar]
- 10.Qin J, Wheeler AR. Lab Chip. 2007;7:186–192. doi: 10.1039/b613414a. [DOI] [PubMed] [Google Scholar]
- 11.Duffy DC, McDonald JC, Schueller OJA, Whitesides G. Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 12.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake S. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy D, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. Nat Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 14.Melin J, Quake S. Annu Rev Biophys Biomol Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 16.Yanik MF, Cinar H, Cinar HN, Chisholm A, Jin Y, Ben-Yakar A. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- 17.Yanik MF, Cinar H, Cinar HN, Chisholm A, Jin Y, Ben-Yakar A. IEEE J Quant Electron. 2006;12:1283–1291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.