Abstract
In Drosophila, detection of tastants is thought to be mediated by members of a family of 68 gustatory receptors (Grs). However, only one receptor, Gr5a, has been associated with a sugar, and it appears to be activated specifically by trehalose. It is unclear whether other sugar receptors are activated by single or multiple sugars. Currently, no Grs are known to colocalize with Gr5a. Such Grs would be candidate sugar receptors because Gr5a-expressing cells function in the responses to attractive tastants. Here we use an “mRNA tagging” approach to identify Gr RNAs that are coexpressed with Gr5a. We found that all seven Grs most related to Gr5a (Gr64a-f and Gr61a) were expressed in Gr5a-expressing cells, whereas none of the other Grs examined were enriched in these Gr neurons (GRNs). We characterized the role of one Gr5a-related receptor, Gr64a, and found that it was required for the behavioral responses to glucose, sucrose, and maltose. Gr64a was required for GRN function because action potentials induced by these sugars were dependent on expression of Gr64a in GRNs. These data demonstrate that multiple Grs are coexpressed with Gr5a and that Drosophila Gr64a is required for the responses to multiple sugars.
Keywords: gustatory receptor neuron, taste, tip recordings, sugars, behavior
The ability to sense sweetness is fundamental to the survival of animals, ranging from flies to humans, because it facilitates the identification of edible, nutrient-rich sources. In Drosophila, detection of sweet, bitter, salt, and pH occurs in gustatory receptor (Gr) neurons (GRNs), rather than neuroepithelial cells, as in mammals (1–3). GRNs are housed in hair-like structures, referred to as sensilla, distributed at the tip of the proboscis (labella), legs, wing margins, and the female genitalia (2).
Drosophila has emerged as an excellent animal model to characterize the sensation of taste. Many studies have focused on the development, distribution, and fine structure of the gustatory organs (2). Moreover, there is recent progress describing the projection patterns of the GRN axons into the brain (4, 5). However, the mechanisms underlying the detection and transduction of tastant-induced stimuli, such as those involved in the sensation of sweetness, are poorly understood.
In mammals the ≈45 Grs fall into two families (T1R and T2R). Multiple T2Rs have been shown to function as bitter receptors, leading to the proposal that all T2Rs may be homomeric bitter receptors (6–13). In contrast, the umami response appears to be mediated by a T1R1/T1R3 heteromer (7, 14–16). Surprisingly, two receptors, T1R2 and T1R3, may account for all responses to sugars through distinct combinations of T1R2/T1R3 heteromers and/or T1R2 and T1R3 homomultimers (7, 14–20). In Drosophila there is a single family of 68 Grs (21–24), and the proportion devoted to sweet, as opposed to bitter, tastants is unresolved. Currently, only two Drosophila Grs have been associated with specific tastants. These include Gr66a, which is required in vivo for the avoidance behavior to caffeine (25), and Gr5a, which has been reported to respond specifically to trehalose (26–28).
Expression of Gr66a and Gr5a reporters indicates that these two Grs are expressed in nonoverlapping subsets of GRNs and appear to define most, but not all, of the GRNs that respond to avoidance and attractive compounds, respectively (4, 5). At least nine additional Grs are expressed in the labellum, and, based on transgenic reporters, all are coexpressed in subsets of the Gr66a-positive GRNs (4, 5). Thus, as in mammals (3), it appears that the majority of Grs participate in the perception of aversive compounds.
In addition to Gr5a, there must be additional sugar-stimulated Grs because Gr5a has been reported to be tuned specifically to trehalose and Gr5a mutant flies display normal behavioral and electrophysiology responses to sucrose (26–28). Other sugar receptors would appear to be coexpressed with Gr5a because Gr5a cells respond to multiple sugars in addition to trehalose (29). However, it is also unclear whether other sugar-activated Grs are activated by a single ligand as appears to be the case for Gr5a.
Here we used a rapid screening approach to identify other Grs that may be coexpressed with Gr5a. We found that the RNAs encoding all seven Grs, which were most related phylogenetically to Gr5a (Gr64a-f and Gr61a), were coexpressed with the Gr5a GRNs. Because these Gr5a-expressing GRNs function in the response to sweet but not bitter tastants, those receptors that are coexpressed with Gr5a would be candidate sugar receptors. We generated Gr64a-deficient flies and showed that this receptor is required for the attractive behavioral responses to glucose, which is the most widely distributed sugar in plants and animals, as well as to the disaccharides, sucrose, and maltose. We also showed that flies missing Gr64a do not generate action potentials in response to these sugars. These results indicate that multiple Grs are coexpressed with Gr5a and that the one receptor/one ligand paradigm for Gr5a does not apply to all sugar receptors in Drosophila.
Results
mRNA Tagging Approach for Identification of Candidate Sugar Receptors.
To identify Grs that were candidate sugar receptors, we tested whether any Gr RNAs were coexpressed with Gr5a. Given that in situ hybridizations to Gr RNAs have been problematic for most Gr genes (4, 5), we used an mRNA tagging approach (30) in combination with the GAL4/UAS system (31). The mRNA tagging approach entails purification of RNAs from specific cell populations using a FLAG-tagged poly(A)-binding protein (PABP). To screen for Grs expressed in Gr5a GRNs, we prepared extracts from dissected labella expressing Gr5a-GAL4;UAS-PABP or Gr66a-GAL4;UAS-PABP transgenes, then immunoprecipated the FLAG-PABP with anti-FLAG antibodies and isolated the mRNAs that were pulled down with the PABP (see Methods).
To test the efficacy of the mRNA tagging, we performed RT-PCR using primers specific for Gr5a and Gr66a. As a control, we performed RT-PCR using primers specific for a pan-neuronally expressed gene, elav (32), which was therefore detected in both Gr5a- and Gr66a-expressing GRNs (Fig. 1A). In contrast to these results, the Gr5a signal was much higher using RNA prepared from Gr5a-GAL4;UAS-PABP than from Gr66a-GAL4;UAS-PABP flies (Fig. 1A). Conversely, the Gr66a product was found primarily by using RNA from Gr66a-GAL4;UAS-PABP flies (Fig. 1A). These results indicated that the mRNA tagging approach may be an effective assay to examine whether other Grs are expressed in either Gr5a- or Gr66a-expressing GRNs.
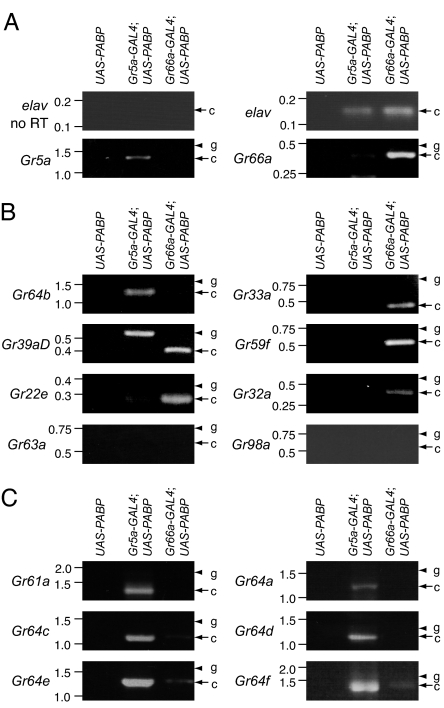
Fig. 1.
RT-PCR screen for Gr RNAs expressed in Gr5a-positive GRNs. (A) Examination of Gr5a and Gr66a RT-PCR products in flies expressing UAS-PABP under control of the Gr5a-GAL4 or Gr66a-GAL4. The RNAs were prepared from the labella of flies containing the UAS-PABP transgene only and from Gr5a-GAL4;UAS-PABP and Gr66a-GAL4;UAS-PABP flies. elav was used as an internal control. The arrows and arrowheads indicate the products generated from the reverse-transcribed mRNA and from the genomic DNA templates, respectively. The sizes of DNA markers (kb) are indicated to the left. (B) RT-PCR products for Gr64b, Gr33a, Gr39aD, Gr59f, Gr22e, Gr32a, Gr63a, and Gr98a. No RT-PCR products for Gr63a or Gr98a were detected. (C) RT-PCR products for the Gr-S group (Gr64 cluster genes and Gr61a).
Initially we surveyed the expression of a set of eight Grs that were distributed among a variety of branches within the Gr family tree [supporting information (SI) Fig. 5] (22). Two of the Gr RNAs (Gr22e and Gr32a) were predicted to be enriched in Gr66a-positive neurons because the corresponding GAL4 reporters have been shown to be expressed in subsets of Gr66a GRNs (4, 5). In addition, we examined Gr63a expression, which was unlikely to be coexpressed with either Gr5a or Gr66a because this Gr encodes a CO2 receptor (33, 34). Gr64b belonged to a distinct branch that included the seven Grs most related to Gr5a (28–45% amino acid identities; referred to here as the Gr-S group).
We found that Gr64b was the only one among the eight surveyed that was enriched in Gr5a-GAL4;UAS-PABP flies (Fig. 1B). In contrast, five of the Gr RNAs were found primarily in the RNA prepared from Gr66a-GAL4;UAS-PABP (Fig. 1B), including three whose expression had not been previously characterized (Gr33a, Gr39aD, and Gr59f). No Gr63a product was detected in either RNA sample (Fig. 1B), which was expected because Gr63a is a CO2 receptor. A Gr98a RT-PCR band also was not detected, which might be because of its low expression level.
Because Gr64b was among the group of seven Grs most related to Gr5a, we tested whether the remaining six members of the Gr-S group were enriched in Gr5a-GAL4;UAS-PABP flies. We found that the RT-PCR products of all Gr-S RNAs (Gr61a and Gr64a-f) were expressed predominately in Gr5a-GAL4;UAS-PABP flies (Fig. 1C). These included Gr64a and Gr64e, despite the report that Gr64a- and Gr64e-GAL4 reporter expression was not detected in the labellum (4). Thus, of the 14 Grs tested, all seven Gr-S RNAs but none of the other Grs were enriched in Gr5a-GAL4;UAS-PABP flies.
Gr64a Is Required for the Behavioral Responses to Multiple Sugars.
We queried the available P-element collections for insertions that disrupted Gr61a or one of the Gr64a-f genes. One P-element was available (GE24923; Genexel, Daejeon, South Korea), which inserted 336 bp upstream from the 5′ end of Gr64a (Fig. 2A). Flies homozygous for the GE24923 insertion were viable and fertile and showed normal responses to all sugar tested and normal levels of the Gr64a RT-PCR product (data not shown). The flies also showed no significant differences from wild-type flies (Canton S) in their sugar preferences (Fig. 3A and SI Fig. 6).
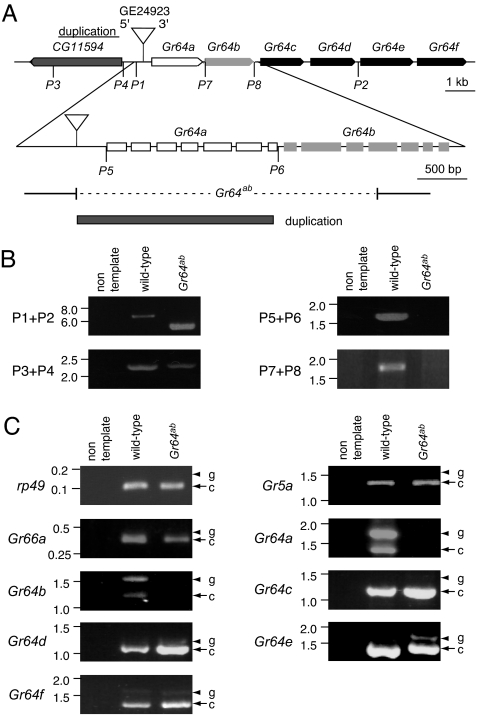
Fig. 2.
Generation of the Gr64ab mutant. (A) The genomic region encoding Gr64a-f. The top horizontal line indicates the portion of the 64A4 region that includes Gr64a-f and CG11594. The pointed end indicates the 3′ end of each gene. The inverted triangle indicates the P-element (GE24923) insertion site. P1 and P2 indicate the locations of the PCR primers used in the primary screen for a deletion flanking GE24923. P3–P8 indicate the primers used in the subsequent analyses. The introns/exons of Gr64a and Gr64b are shown at a higher resolution below. One line (Gr64ab) contained a 3.1-kb deletion, which extended from the P-element insertion site to the fourth exon of Gr64b (dashed line), and a duplication of a 2.0-kb portion of CG11594 as indicated. (B) PCR analyses of the Gr64ab region. The PCR product using the P1 and P2 primers generated 6.2- and 5.0-kb products from wild type and Gr64ab, respectively. The P3 and P4 primers, which flank the duplicated region, produce the same products in wild type and Gr64ab. The P5/P6 and P7/P8 primer pairs produced products only in wild type because of the deletion in Gr64ab. The precise alterations were determined subsequently by DNA sequencing. (C) Analyses of Gr64a-f RT-PCR products. The control RT-PCR products (rp49, Gr5a, and Gr66a) were not affected in Gr64ab. The arrows and arrowheads indicate the products generated from the reverse-transcribed mRNA and from the genomic DNA templates, respectively. DNA size markers (kb) are shown.
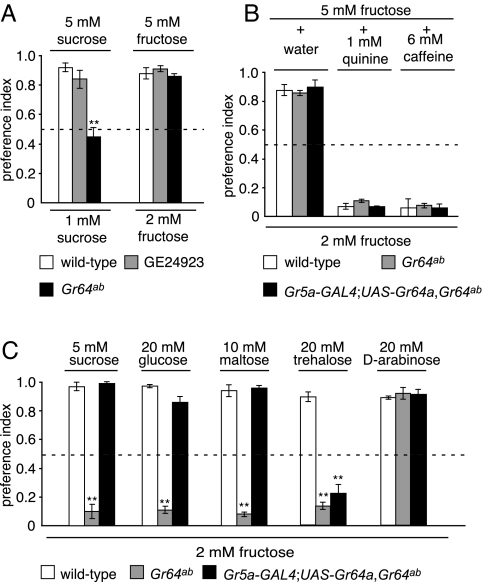
Fig. 3.
Two-way choice tests showing that Gr64a was required for the behavioral responses to multiple sugars. PI values of 1.0 or 0 indicate complete preferences for one or the other tastant, and a PI of 0.5 indicates a lack of preference. (A) Flies were allowed to choose between two concentrations of the same sugar (sucrose or fructose). (B) Gr64a was not required for the avoidance of quinine or caffeine. The flies were given the choice between 2 mM fructose or 5 mM fructose plus either 1 mM quinine or 6 mM caffeine. (C) Two-sugar competition assay. The following fly stocks were allowed to choose between 2 mM fructose and higher concentrations of other sugars as indicated: (i) wild type (Canton S), (ii) Gr64ab, and (iii) Gr64ab flies containing the Gr5a-GAL4;UAS-Gr64a transgenes. The dashed line indicates a PI of 0.5. Statistically significant differences from wild type were checked by using the unpaired Student t test (**, P < 0.01).
To generate a deletion flanking the 3′ end of GE24923, we genetically introduced the transposase because mobilization of P-elements can result in deletions flanking the site of the original insertion. After screening genomic DNA from ≈800 lines by PCR, we identified one with a 3.1-kb deletion, which removed all of Gr64a and 868 bp at the 5′ end of Gr64b. In addition, a 2.0-kb portion of the flanking gene, CG11594, was duplicated and inserted in the deleted region (Fig. 2A). The original CG11594 gene was not disrupted by the deletion (Fig. 2B). This mutant line is referred to as Gr64ab.
We examined whether the Gr64 cluster genes were expressed in Gr64ab homozygous flies by performing RT-PCR using total RNA prepared from labella. We found that expression of the Gr64a and Gr64b mRNAs was eliminated, whereas the mRNAs from all of the other four genes (Gr64c, Gr64d, Gr64e, and Gr64f) in the cluster were still produced (Fig. 2C).
To determine whether Gr64ab flies had a defect in detecting sugars, we used a two-sugar choice test. When presented with a choice between 5 mM sucrose and 2 mM fructose, wild-type flies displayed a very strong preference to consume the sucrose (Fig. 3C) [preference index (PI) = 0.97 ± 0.03; n = 6]. This bias for selecting sucrose was not due to an inability to detect fructose, because wild-type flies selected the higher of two concentrations of fructose (5 mM versus 2 mM) (Fig. 3A) (PI = 0.88 ± 0.04; n = 3), similar to the situation when offered two different concentrations of sucrose (5 mM versus 1 mM) (Fig. 3A) (PI = 0.92 ± 0.03; n = 5).
In contrast, Gr64ab flies preferred fructose to sucrose (Fig. 3C) (PI = 0.10 ± 0.05; n = 6). This suggested that the response to sucrose was reduced or eliminated. In addition, Gr64ab flies failed to show a bias in favor of the 5 mM over the 1 mM sucrose (Fig. 3A) (PI = 0.45 ± 0.06; n = 5) but retained the ability to select the 5 mM over 2 mM fructose (Fig. 3A) (PI = 0.86 ± 0.02; n = 3). Thus, the behavioral response to sucrose, but not fructose, was defective in Gr64ab flies.
We used the two-sugar choice assay to test the responses of Gr64ab flies to other sugars. In the case of wild-type flies, there was a strong propensity to consume 20 mM glucose, 10 mM maltose, 20 mM trehalose, or 20 mM arabinose over 2 mM fructose (Fig. 3C) (PI = 0.98 ± 0.01, 0.94 ± 0.04, 0.89 ± 0.03, and 0.89 ± 0.01, respectively; n = 3–6). The Gr64ab flies also exhibited a normal preference for 20 mM arabinose over 2 mM fructose (Fig. 3C) (PI = 0.92 ± 0.04; n = 3). However, unlike wild type, the Gr64ab mutant preferred fructose over either glucose or maltose (Fig. 3C) (PI = 0.11 ± 0.03 and 0.08 ± 0.02, respectively; n = 6). Also, in contrast to wild type, Gr64ab selected fructose rather than trehalose (Fig. 3C) (PI = 0.15 ± 0.02; n = 6), which was surprising given that trehalose is the ligand that activates Gr5a (26–28).
Bitter compounds, such as quinine and caffeine, are aversive to wild-type flies, and this response is mediated by GRNs that express Gr66a but not Gr5a (4, 5). Consistent with the expression of the Gr64a and Gr64b RNAs in Gr5a-expressing GRNs, we found that Gr64ab flies showed the normal aversion to quinine and caffeine (Fig. 3B) (wild type, PI = 0.07 ± 0.02 and 0.06 ± 0.03, respectively; Gr64ab, PI = 0.11 ± 0.01 and 0.08 ± 0.02; n = 3, respectively).
Because both Gr64a and Gr64b were deleted in the Gr64ab mutant, we addressed whether the defects in the behavioral responses to sucrose, glucose, maltose, and trehalose detection were due to loss of Gr64a or Gr64b. We generated UAS-Gr64a and UAS-Gr64b transgenes and expressed them in Gr64ab flies under the control of the Gr5a-GAL4. The UAS-Gr64b and Gr5a-GAL4 transgenes did not restore a normal response to any sugar, including trehalose (data not shown). However, introduction of the UAS-Gr64a transgene, in combination with the Gr5a-GAL4, fully restored the preferences for sucrose, glucose, or maltose over fructose (Fig. 3C) (PI = 0.99 ± 0.01, 0.86 ± 0.04, and 0.96 ± 0.02, respectively; n = 6) but not the trehalose response (PI = 0.22 ± 0.06; n = 6). Introduction of both UAS-Gr64a and UAS-Gr64b with Gr5a-GAL4 also did not rescue the trehalose response (data not shown). Nevertheless, the trehalose defect appeared to be associated with the Gr64ab mutation, rather than a background mutation, because the phenotype was observed in flies containing the Gr64ab chromosome in trans with either of two deficiency chromosomes that span the Gr64 locus (SI Fig. 7). Furthermore, introduction of a transgene that included the entire CG11594 genomic region did not rescue any aspect of the Gr64ab phenotype (data not shown). These results demonstrate that Gr64a functions in the detection of sucrose, glucose, and maltose in Gr5a-expressing GRNs. As with many Grs, we were unable to detect expression of Gr64a RNA by in situ hybridizations, presumably because of low expression levels.
Sucrose-, Glucose-, and Maltose-Induced Action Potentials Require Gr64a.
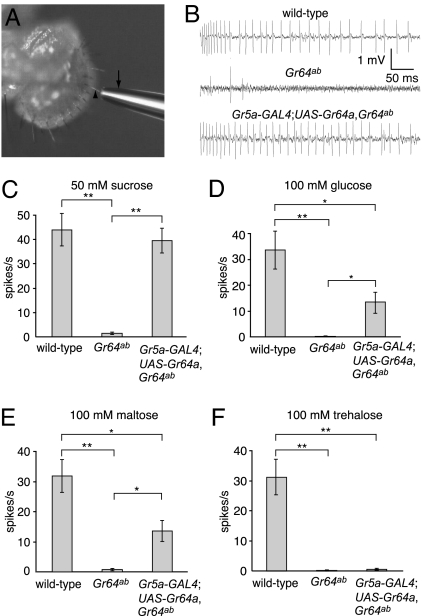
To address whether the electrophysiological response to sugars was defective in Gr64a-deficient flies, we assayed tastant-induced action potentials in the GRNs by performing tip recordings (Fig. 4A). In Drosophila, sugar-induced action potentials can be detected in the GRNs in L, I, or S type bristles, although the highest frequencies and rates of response occur in L type sensilla (35). Therefore, we applied sugars to L type bristles and compared the frequencies of action potentials in wild type and in the Gr64ab mutant. In wild-type flies, sucrose, glucose, maltose, and trehalose stimulated high frequencies of action potentials (Fig. 4 B–F) (44.0 ± 6.7, 34.4 ± 8.7, 31.8 ± 5.5, and 33.4 ± 5.7, respectively; n = 5–7), whereas the frequencies of action potentials induced by these sugars in Gr64ab were dramatically reduced (Fig. 4 B–F) (1.4 ± 0.5, 0.2 ± 0.2, 0.8 ± 0.3, and 0.8 ± 0.5, respectively; n = 5–7). Consistent with the behavioral assays, the reduction in action potentials in response to trehalose was not rescued by either of the wild-type Gr64a or Gr64b transgenes (Fig. 4F) (0.6 ± 0.04; n = 5 and data not shown). In contrast, introduction of the Gr5a-GAL4 and UAS-Gr64a transgenes restored action potentials to various extents in response to sucrose, glucose, and maltose (Fig. 4 B–E) (39.6 ± 5.0, 12.8 ± 4.9, and 13.6 ± 3.4, respectively; n = 5–7). These results indicated that Gr64a was required in GRNs for the detection of sucrose, glucose, and maltose.
Fig. 4.
Gr64a was required for sucrose-, glucose-, and maltose-induced action potentials. Tip recordings were performed on L4 sensilla of the indicated genotypes. (A) Recording electrode (arrow) placed over an l type bristle (arrowhead). (B) Sample tip recordings using 50 mM sucrose. (C–F) Average frequencies of action potentials (spikes per second) in response to the indicated sugars. The averages were based on data collected between 50 msec and 1,050 msec after application of the sugars (n = 5–7). Statistically significant differences between the indicated pairs of data were checked by using the unpaired Student t test (*, P < 0.05; **, P < 0.01).
Discussion
Before the current analysis, the only known Drosophila Grs linked to specific tastants were Gr5a (26–28) and Gr66a (25), which are required for the responses to trehalose and caffeine, respectively. All nine of the other Grs shown to be expressed in the labellum appeared to be coexpressed in subsets of Gr66a-expressing GRNs (4, 5). Thus, there were many candidate bitter Grs. However, the identities of candidate sugar-responsive Grs, or whether any Grs were coexpressed with the trehalose receptor, were not known. An additional question is whether other sugar receptors were tuned to single or multiple sugars.
Because in most cases detection of Gr RNAs by in situ hybridizations has been unsuccessful, we screened for Grs that are coexpressed with Gr5a using an mRNA tagging approach. We found that expression of the seven Grs that were most related to Gr5a (Gr64a-f and Gr61a) were enriched in Gr5a-expressing GRNs. Thus, all eight members of this phylogenetically distinct group, referred to as Gr-S, are candidate sugar receptors. None of the other 14 Grs analyzed here or in previous studies are coexpressed with Gr5a (4, 5). Because T1R2 and T1R3 seem to account for all of the sugar responses in mammals (16, 36), it may be that there is a larger repertoire of sugar receptors in Drosophila.
To test the proposal that Gr-S receptors other than Gr5a are sugar receptors, we examined the requirement for Gr64a. An additional question is whether other Drosophila sugar receptors are activated by one or multiple sugars. In contrast to the mammalian sugar receptors, Gr5a was reported to be specifically activated by trehalose (26–28). This observation, in combination with the larger number of candidate sugar receptors in flies than mammals, raised the possibility that each Gr-S member may respond primarily to one sugar. However, we found that the Gr64a gene was required in vivo for the responses to the monosaccharide, glucose, and the disaccharides, sucrose and maltose, each of which includes at least one glucose subunit. Gr64a was not essential for the responses to all sugars, because the Gr64ab flies responded normally to the monosaccharides, fructose and arabinose. Although the defects in the behavioral responses to sucrose, glucose, and maltose were completely rescued by expression of a wild-type Gr64a transgene, only the sucrose response was fully rescued as assayed by tip recordings. The rescues of the electrophysiological responses to glucose and maltose were significant but did not restore the same frequencies of action potentials as in wild type. Although the explanation for this result is unclear, similar findings were reported for rescue of the trehalose deficits in the Gr5aΔ5 allele by a wild-type transgene (28). Whereas the behavioral phenotype in Gr5aΔ5 was restored entirely, the rescue of the electrophysiological response to trehalose was partial.
The trehalose response was also greatly reduced in Gr64ab mutant flies. This result was unexpected because the response to this sugar is nearly eliminated in the Gr5a mutant, and Gr5a is sufficient to confer trehalose sensitivity in S2 cells (26). Nevertheless, there is small residual trehalose response in the Gr5a mutant (28). The trehalose defect in Gr64ab flies did not appear to arise from a background mutation in Gr5a, because the phenotype was observed in flies in which the Gr64ab deletion was placed in trans with deficiencies that spanned the Gr64 locus. Thus, the question arises as to the identity of the second trehalose receptor. The deletion in Gr64ab disrupts both Gr64a and Gr64b, and introduction of a wild-type UAS-Gr64a transgene under the control of the Gr5a-GAL4 restores normal responses to sucrose, glucose, and maltose, but not to trehalose. Thus, Gr64b may be a trehalose receptor. However, a UAS-Gr64b transgene alone or in combination with the UAS-Gr64a failed to restore a trehalose response in flies containing the Gr5a-GAL4 (Y.J. and C.M., unpublished data), suggesting that either Gr64b is not a trehalose receptor or the transgene is nonfunctional.
A general issue concerning the Drosophila Grs is whether they typically form homo- or heteromultimers. An indication that at least some Grs form obligatory heteromultimers is that misexpression of just one of the two CO2 receptor genes, Gr21a or Gr63a, in CO2-insensitive antennal neurons is insufficient to confer CO2 sensitivity to these cells. However, coexpression of both Gr21a and Gr63a induces CO2 responsiveness (33, 34). Misexpression of just Gr64a in Gr66a GRNs did not elicit an aversive response to sucrose, glucose, or maltose or result in sugar-induced action potentials in Gr66a GRNs (Y.J. and C.M., unpublished data). Similarly, expression of the caffeine receptor Gr66a in Gr5a-expressing cells does not confer caffeine sensitivity to these cells (S.J.M. and C.M., unpublished data). These results raise the possibility that these and possibly other taste receptors in flies are obligatory heterodimers, as is the case for the CO2 receptors. The other Gr-S members would appear to be the best candidates for forming heteromultimers with Gr64a. Finally, we propose that the mRNA tagging approach applied here can be extended to identify pairs of Grs that are expressed together in smaller subsets of GRNs and would therefore be excellent candidates for forming heteromultimers.
Methods
Genetics, Fly Stocks, and Constructs.
The GE24923 P-element (Genexel), which inserted 366 bp 5′ of Gr64a, was mobilized by genetically introducing the transposase using the Δ2-3 line (37): w;Sp/CyO; ry Sb1 P{ry[+t7.2] = Δ2-3}99B/TM6B (Bloomington Drosophila Stock Center, Bloomington, IN). To identify the Gr64ab deletion, we screened ≈800 lines by PCR as previously described (38) using the following primers: P1, 5′-TTATTAGAAGCGCGCACACCTA CTC-3′; P2, 5′-ACAAGGATATCCAGCGAAAGCGCA-3′ (Fig. 2A).
To create the P[UAS-Gr64a] and P[UAS-Gr64b] transgenes, we amplified the coding regions of Gr64a and Gr64b from total labellar RNA by RT-PCR and subcloned the cDNAs into the pUAST vector (31). The clones were sequenced, confirming that no mutations were introduced. The transgenic lines were generated by using standard procedures, and the transgenes were crossed into the w1118;Gr64ab background. The P[Gr5a-GAL4] and P[Gr66a-GAL4] transgenic flies were kindly provided by H. Amrein (Duke University Medical Center, Durham, NC).
Chemicals.
Sucrose, glucose, maltose, d-arabinose, quinine, caffeine, and tricholine citrate were purchased from Sigma-Aldrich (St. Louis, MO), and trehalose was from Fluka (St. Gallen, Switzerland).
mRNA Tagging.
The mRNA tagging protocol was modified from that described previously (30). Approximately 400 fly labella were dissected and fixed in 1 ml of PBS with 1% formaldehyde and 0.5% Nonidet P-40 for 30 min at 4°C. A total of 140 μl of 2 M glycine was added, and the samples were incubated for an additional 5 min at 4°C. The samples were washed three times with 1 ml of PBS and homogenized in 0.8 ml of homogenization buffer (HB: 150 mM NaCl/50 mM Hepes, pH 7.6/1 mM EGTA/15 mM EDTA/10% glycerol). Immediately before addition of the HB, we added the following to the HB at the indicated final concentrations: 8 mM vanadyl ribonucleoside complex (Sigma-Aldrich), 50 units/ml SUPERase-In (Ambion, Austin, TX), and a protease inhibitor mixture tablet (one tablet per 50 ml; Roche, Indianapolis, IN). The homogenate was then sonicated for 1 min at 30% intensity (using a Fisher Sonic Dismembrator, Model 500) and cleared by centrifugation for 10 min at 13,000 × g. Anti-FLAG-M2 affinity agarose beads (Sigma-Aldrich) were washed four times with HB at 4°C by centrifugation for 1 min at 1,500 × g. To coimmunoprecipitate the FLAG-tagged PABP and the associated polyadenylated mRNAs, we added 100 μl of anti-FLAG-M2 affinity agarose beads to the cleared homogenate, which was then incubated for 2 h at 4°C. The beads were washed four times with the HB at 4°C and incubated in 100 μl of elution buffer (50 mM Tris·HCl, pH 7.0/10 mM EDTA/1.3% SDS/50 units/ml SUPERase-In) at 65°C for 30 min to reverse the RNA::PABP crosslink. A total of 100 μl of eluant was treated with 400 μl of TRIzol (Invitrogen, Carlsbad, CA) and 100 μl of chloroform, and the RNA extracted was finally dissolved in 40 μl of RNase-free water. The P[UAS-hPABP-FLAG] transgenic flies used for these experiments were kindly provided by R. L. Davis (Baylor College of Medicine, Houston, TX).
RT-PCR Amplification of Gr RNAs.
Two types of RNA preparations were used for performing the RT-PCR: (i) the mRNA tagging approach was used to prepare the RNA as described above (Fig. 1), or (ii) 100 labella were dissected and total RNA was extracted by using TRIzol reagent (Invitrogen) (Fig. 2). The RNA samples were treated with DNaseI (Invitrogen) before the RT-PCRs were performed. Each pair of primers to a given Gr was designed to span at least one intron so that the products derived from mRNA and genomic DNA could be discriminated on the basis of size. Because elav has no introns, we used a no-reverse-transcriptase control to confirm that the RT-PCR product was derived from mRNA instead of genomic DNA. A OneStep RT-PCR Kit (catalog no. 210212; Qiagen, Valencia, CA) was used for the RT-PCR. To perform the RT-PCR with total RNA we amplified for 30–35 cycles, and for the mRNA tagging experiments we used 40 cycles.
Behavioral Assays.
The two-way choice assays were performed by using modifications of previously described procedures (25, 39). For each assay, 30 flies (3–6 days old) were starved overnight on 1% agarose and then introduced into 72-well microtiter dishes filled with two types of test mixtures placed in alternating wells. Each test mixture contained 1% agarose and either blue dye (0.125 mg/ml brilliant blue FCF, catalog no. 027-12842; Wako Chemical, Richmond, VA) or red dye (0.2 mg/ml sulforhodamine B, catalog no. S9012; Sigma-Aldrich) and a test tastant. For the attractive tests, the wells were filled with (i) one sugar at two different concentrations or (ii) 2 mM fructose and a different sugar at a higher concentration (5–20 mM). A higher concentration of the second sugar was selected if it induced a PI of >0.8. The avoidance assays were conducted by using 2 mM fructose versus 5 mM fructose plus either 1 mM quinine or 6 mM caffeine. After allowing the flies to feed for 90 min at room temperature in the dark, the animals were frozen in the dishes at −20°C, and the numbers of flies with blue (NB), red (NR), or purple (NP) abdomens were assessed by visual inspection. In those cases in which the colors were difficult to judge, the guts were dissected. If the amount of red dye was between 50% and 150% of the blue dye, the color was scored as purple. If the red dye was >150% or <50% of the blue dye, the fly was counted as red and blue, respectively. All behavioral assays (30 flies per test) were performed three to six times. The PI values were calculated according to the following equation: PI = (NB + 0.5 NP)/NTotal or (NR + 0.5 NP)/NTotal. The dyes did not cause preference changes, because no differences in PIs were caused by switching the dyes. For example, in those assays in which wild-type flies were allowed to choose between 5 mM sucrose and 2 mM fructose, the PI values were 0.96 ± 0.04 when blue and red dyes were used for sucrose and fructose, respectively, and 0.97 ± 0.03 when blue and red dyes were used for fructose and sucrose, respectively. The wild-type control was Canton S, which showed behavioral responses indistinguishable from the parental P-element insertion line (GE24923) (SI Fig. 6).
Electrophysiology.
Tip recordings on labellar bristles were performed according to procedures similar to those previously described (25). Briefly, to provide a reference electrode and to stabilize the fly for the recordings, we inserted a glass capillary with Ringer's solution into the abdomen so that it extended through to the fly head. The electrolyte used in the recording electrode (10–20 μm in diameter) was 30 mM tricholine citrate. The recordings were performed on the L4 sensilla (flies ≤1 day after eclosion). The signals were collected and amplified from the recording electrode through a preamplifier (TastePROBE; Syntech, Hilversum, The Netherlands) and a 100- to 3,000-Hz band-pass filter. Autospike 3.1 software (Syntech) was used to acquire the action potentials (9.6-kHz sampling rate) and to analyze the frequencies. For most sugars we used 100 mM, which is the concentration typically used in previous analyses (28, 35). We used 50 mM sucrose because this sugar induced a much higher frequency of action potentials than other sugars. All recordings using a given genotype and tastant were performed five to seven times.
Data Analyses.
All error bars represent SEMs. Unpaired Student's t tests were used to check for significant differences between the indicated pairs of data (*, P < 0.05; **, P < 0.01).
Supplementary Material
Acknowledgments
We thank Dr. R. L. Davis for the P[UAS-hPABP-FLAG] transgenic flies, Dr. H. Amrein for the P[Gr5a-GAL4] and P[Gr66a-GAL4] transgenic flies, and Dr. T. Wang for comments on the manuscript and assistance with the figures. This work was supported by Grant DC007864 from the National Institute on Deafness and Other Communication Disorders (to C.M.).
Abbreviations
- Gr
gustatory receptor
- GRN
Gr neuron
- PABP
poly(A)-binding protein
- PI
preference index.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702421104/DC1.
References
- 1.Ebbs ML, Amrein H. Pflügers Arch. 2007;454:735–747. doi: 10.1007/s00424-007-0246-y. [DOI] [PubMed] [Google Scholar]
- 2.Stocker RF. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 3.Scott K. Neuron. 2005;48:455–464. doi: 10.1016/j.neuron.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Thorne N, Chromey C, Bray S, Amrein H. Curr Biol. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Singhvi A, Kong P, Scott K. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 7.Matsunami H, Montmayeur JP, Buck LB. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 8.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 9.Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 10.Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 11.Behrens M, Brockhoff A, Kuhn C, Bufe B, Winnig M, Meyerhof W. Biochem Biophys Res Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Proc Natl Acad Sci USA. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. Proc Natl Acad Sci USA. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 17.Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 18.Sainz E, Korley JN, Battey JF, Sullivan SL. J Neurochem. 2001;77:896–903. doi: 10.1046/j.1471-4159.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- 19.Montmayeur JP, Liberles SD, Matsunami H, Buck LB. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- 20.Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 21.Clyne PJ, Warr CG, Carlson JR. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- 22.Robertson HM, Warr CG, Carlson JR. Proc Natl Acad Sci USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott K, Brady R, Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 24.Dunipace L, Meister S, McNealy C, Amrein H. Curr Biol. 2001;11:822–835. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- 25.Moon SJ, Kottgen M, Jiao Y, Xu H, Montell C. Curr Biol. 2006;16:1812–1817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Chyb S, Dahanukar A, Wickens A, Carlson JR. Proc Natl Acad Sci USA. 2003;100:14526–14530. doi: 10.1073/pnas.2135339100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueno K, Ohta M, Morita H, Mikuni Y, Nakajima S, Yamamoto K, Isono K. Curr Biol. 2001;11:1451–1455. doi: 10.1016/s0960-9822(01)00450-x. [DOI] [PubMed] [Google Scholar]
- 28.Dahanukar A, Foster K, van der Goes van Naters WM, Carlson JR. Nat Neurosci. 2001;4:1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- 29.Marella S, Fischler W, Kong P, Asgarian S, Rueckert E, Scott K. Neuron. 2006;49:285–295. doi: 10.1016/j.neuron.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Edenberg HJ, Davis RL. Nucleic Acids Res. 2005;33:e148. doi: 10.1093/nar/gni149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand AH, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 32.Robinow S, White K. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- 33.Kwon JY, Dahanukar A, Weiss LA, Carlson JR. Proc Natl Acad Sci USA. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones WD, Cayirlioglu P, Grunwald Kadow I, Vosshall LB. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 35.Hiroi M, Marion-Poll F, Tanimura T. Zool Sci. 2002;19:1009–1018. doi: 10.2108/zsj.19.1009. [DOI] [PubMed] [Google Scholar]
- 36.Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Varadarajan V, Zou S, Jiang P, Ninomiya Y, Margolskee RF. Science. 2003;301:850–853. doi: 10.1126/science.1087155. [DOI] [PubMed] [Google Scholar]
- 37.Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Lee SJ, Suzuki E, Dugan KD, Stoddard A, Li HS, Chodosh LA, Montell C. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanimura T, Isono K, Takamura T, Shimada I. J Comp Physiol A. 1982;147:433–437. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.