Abstract
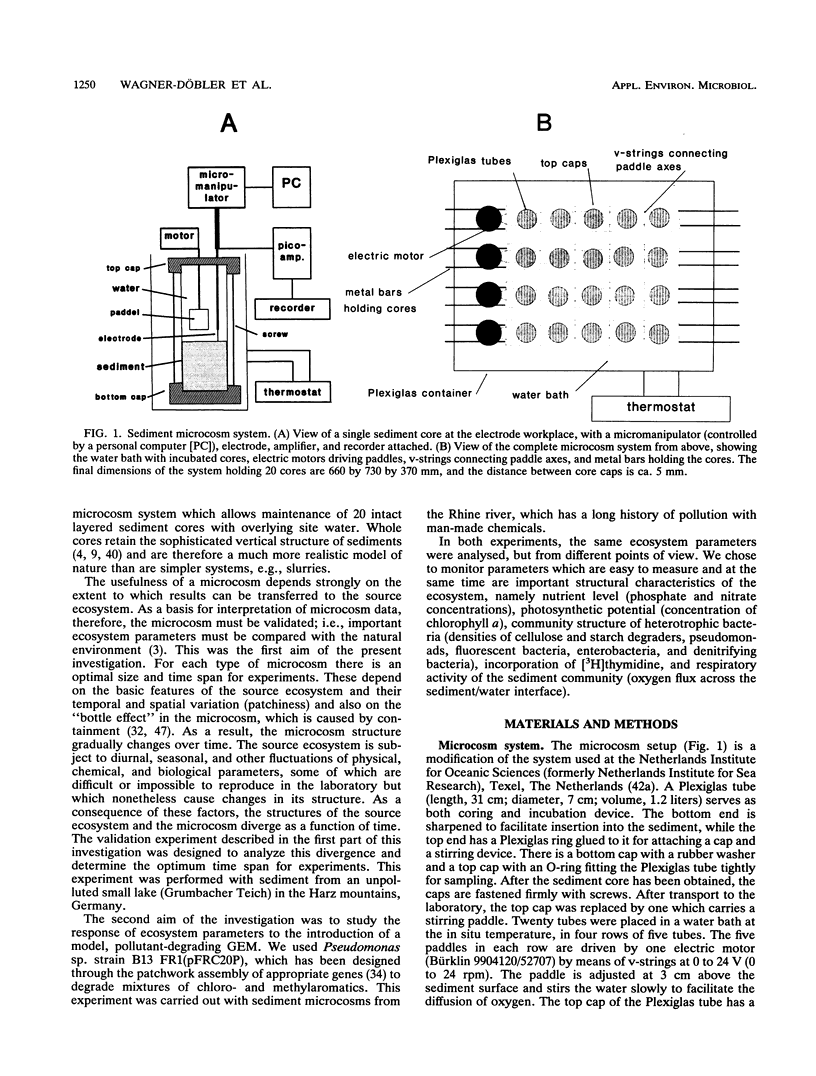
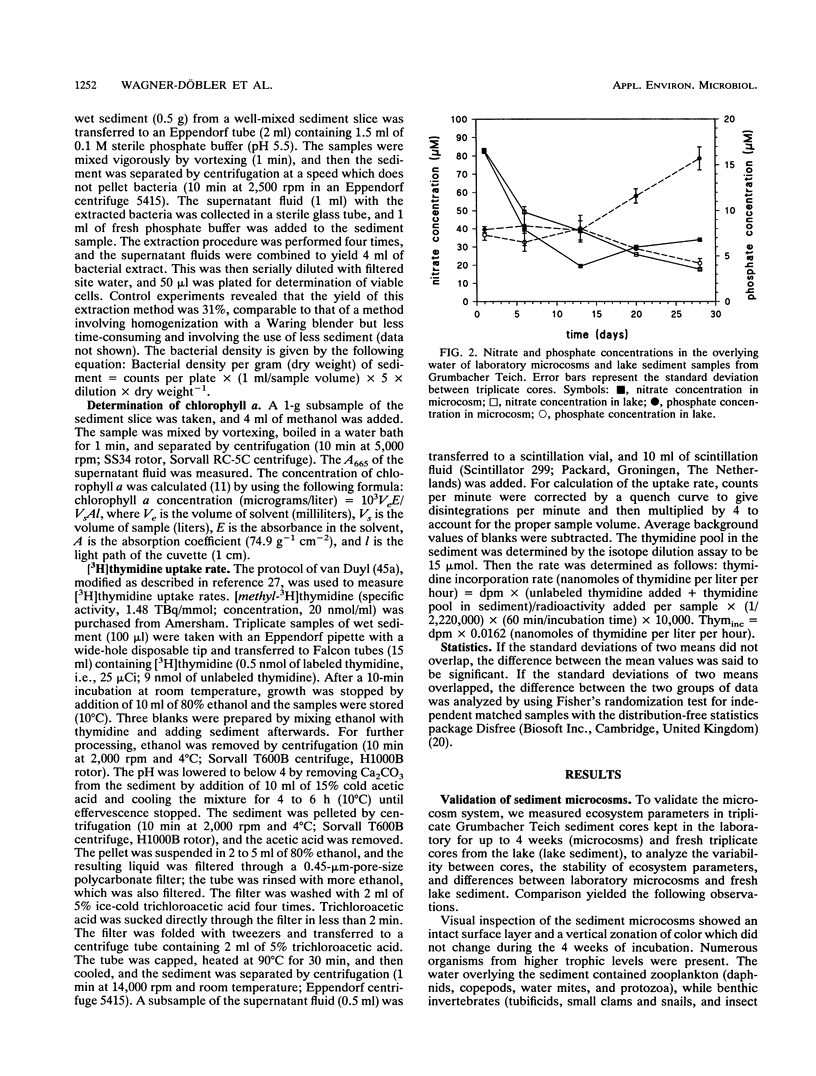
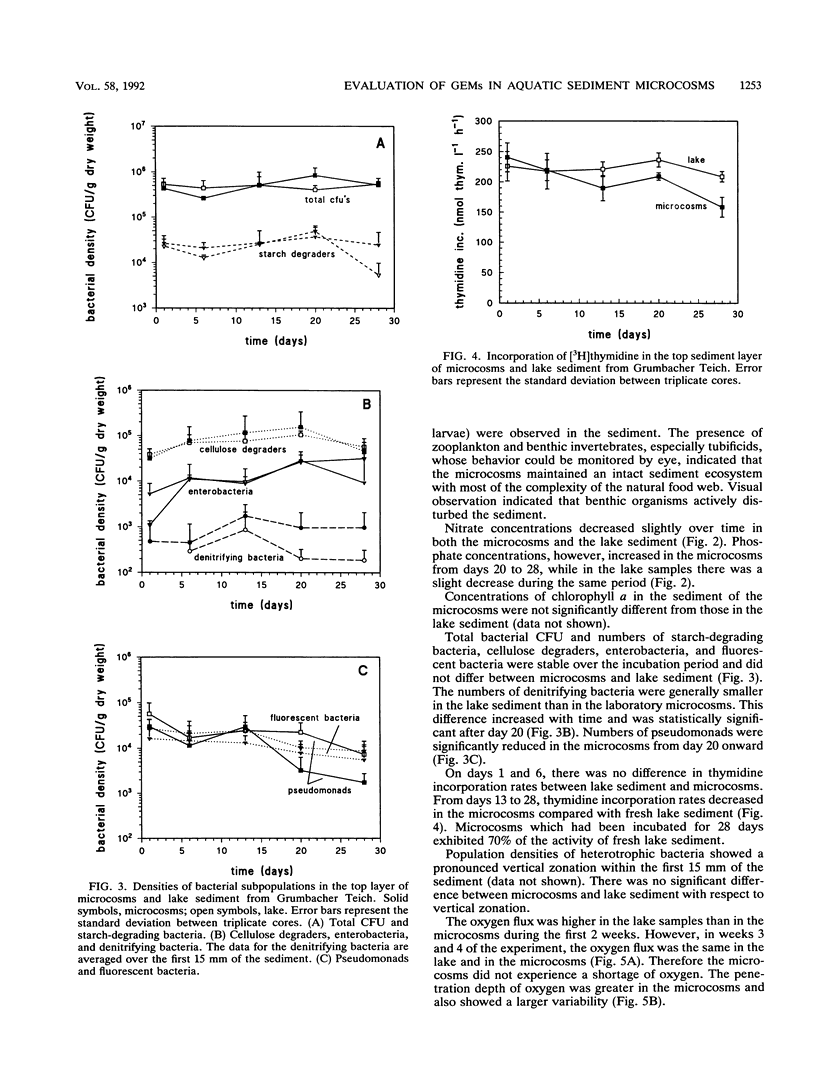
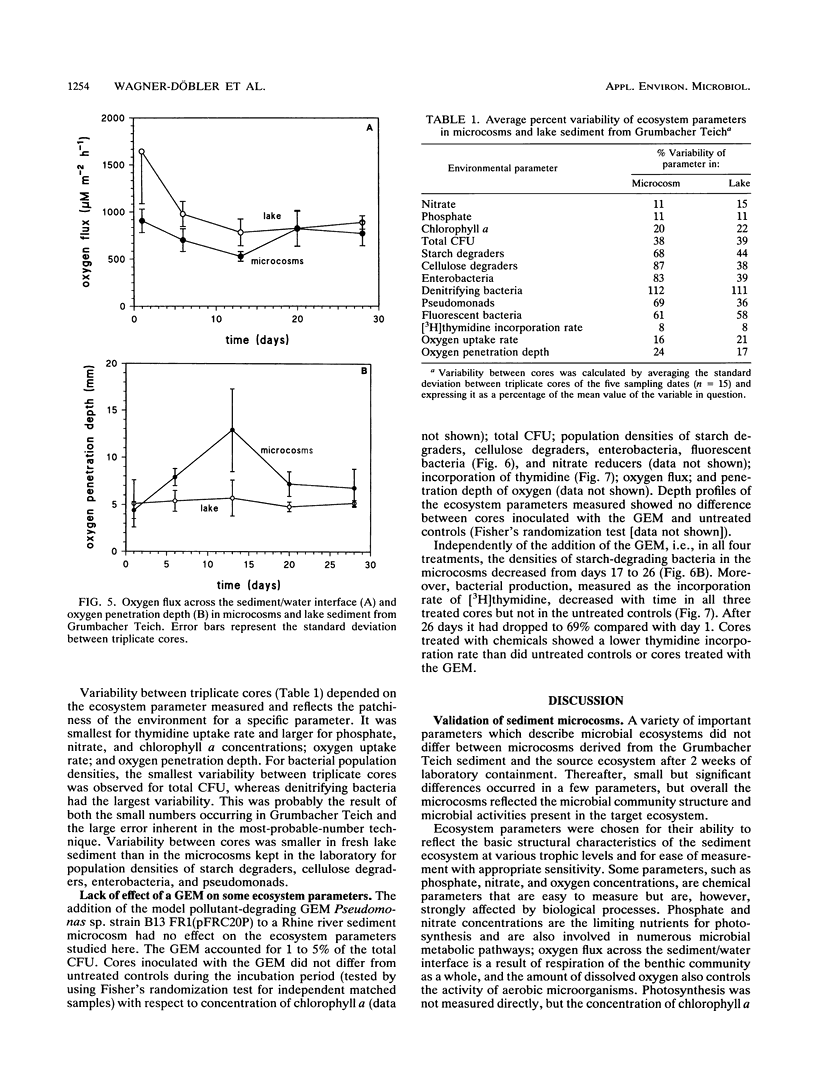
In this paper we describe a sediment microcosm system consisting of 20 undisturbed, layered sediment cores with overlying site water which are incubated under identical conditions of temperature, light, stirring rate of overlying water, and water exchange rate. Ecosystem parameters (nutrient level, photosynthetic potential, community structure of heterotrophic bacteria, thymidine incorporation rate, and oxygen microgradients) of the laboratory microcosms and the source ecosystem were compared and shown to be indistinguishable for the first 2 weeks. In weeks 3 and 4, small differences were detectable in the nutrient level, community structure of heterotrophic bacteria, and thymidine incorporation rate. However, the photosynthetic potential, depth profiles of heterotrophic bacterial community structure, and oxygen microgradients were maintained throughout the incubation period and did not differ between laboratory microcosms and the source ecosystem. The microcosm system described here would thus appear to be a valid model of aquatic sediments for up to 4 weeks; the actual period would depend on the sediment source and incubation temperature. The validated systems were used with Rhine river sediment to assess possible effects on ecosystem parameters of Pseudomonas sp. strain B13 FR1(pFRC20P), a genetically engineered microorganism (GEM) that had been constructed to degrade mixtures of halo- and alkylbenzoates and -phenols. The GEM survived in the surface sediment at densities of 5 x 10(4) to 5 x 10(5)/g (dry weight) for 4 weeks and degraded added chloro- and methylaromatics. The GEM did not measurably influence ecosystem parameters such as photosynthesis, densities of selected heterotrophic bacteria, thymidine incorporation rate, and oxygen microgradients. Thus, the microcosm system described here would seem to be useful for the study of the ecology of biodegradation and the fate and effect of microorganisms introduced into the environment.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander M. Biodegradation of chemicals of environmental concern. Science. 1981 Jan 9;211(4478):132–138. doi: 10.1126/science.7444456. [DOI] [PubMed] [Google Scholar]
- Awong J., Bitton G., Chaudhry G. R. Microcosm for assessing survival of genetically engineered microorganisms in aquatic environments. Appl Environ Microbiol. 1990 Apr;56(4):977–983. doi: 10.1128/aem.56.4.977-983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P. B., Nielsen L. P., Revsbech N. P., Sørensen J. Microzonation of denitrification activity in stream sediments as studied with a combined oxygen and nitrous oxide microsensor. Appl Environ Microbiol. 1989 May;55(5):1234–1241. doi: 10.1128/aem.55.5.1234-1241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. M., Mallory L. M., Alexander M. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol. 1985 Oct;50(4):977–983. doi: 10.1128/aem.50.4.977-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks C. W. Increased recovery rate of salmonellae from stream bottom sediments versus surface waters. Appl Microbiol. 1971 Feb;21(2):379–380. doi: 10.1128/am.21.2.379-380.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Sayler G. S. Problems and potential for in situ treatment of environmental pollutants by engineered microorganisms. Microbiol Sci. 1987 Feb;4(2):59–63. [PubMed] [Google Scholar]
- Kong H. L., Sayler G. S. Degradation and total mineralization of monohalogenated biphenyls in natural sediment and mixed bacterial culture. Appl Environ Microbiol. 1983 Sep;46(3):666–672. doi: 10.1128/aem.46.3.666-672.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979 Nov 15;100(1):95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Maziarz Z., Gaik A., Szymański A., Zajgner J. Ocena układu oddechowego w najcześciej spotykanych schorzeniach płuc, za pomoca radiograficznej i radioizotopowej metody badania ksenonem 133Xe. Pol Przegl Radiol Med Nukl. 1979 Sep-Oct;43(5):272–273. [PubMed] [Google Scholar]
- McClure N. C., Fry J. C., Weightman A. J. Survival and catabolic activity of natural and genetically engineered bacteria in a laboratory-scale activated-sludge unit. Appl Environ Microbiol. 1991 Feb;57(2):366–373. doi: 10.1128/aem.57.2.366-373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure N. C., Weightman A. J., Fry J. C. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl Environ Microbiol. 1989 Oct;55(10):2627–2634. doi: 10.1128/aem.55.10.2627-2634.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky J. A. Microbial growth rates and biomass production in a marine sediment: evidence for a very active but mostly nongrowing community. Appl Environ Microbiol. 1987 Oct;53(10):2368–2372. doi: 10.1128/aem.53.10.2368-2372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvos D. R., Lacy G. H., Cairns J. Genetically Engineered Erwinia carotovora: Survival, Intraspecific Competition, and Effects upon Selected Bacterial Genera. Appl Environ Microbiol. 1990 Jun;56(6):1689–1694. doi: 10.1128/aem.56.6.1689-1694.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipke R., Wagner-Döbler I., Timmis K. N., Dwyer D. F. Survival and function of a genetically engineered Pseudomonad in aquatic sediment microcosms. Appl Environ Microbiol. 1992 Apr;58(4):1259–1265. doi: 10.1128/aem.58.4.1259-1265.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quensen J. F., 3rd, Tiedje J. M., Boyd S. A. Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediments. Science. 1988 Nov 4;242(4879):752–754. doi: 10.1126/science.242.4879.752. [DOI] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Scanferlato V. S., Orvos D. R., Cairns J., Lacy G. H. Genetically Engineered Erwinia carotovora in Aquatic Microcosms: Survival and Effects on Functional Groups of Indigenous Bacteria. Appl Environ Microbiol. 1989 Jun;55(6):1477–1482. doi: 10.1128/aem.55.6.1477-1482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerman P. R., Schmidt J. P., Alexander M. Factors affecting the survival and growth of bacteria introduced into lake water. Arch Microbiol. 1988;150(4):320–325. doi: 10.1007/BF00408301. [DOI] [PubMed] [Google Scholar]
- Shiaris M. P., Rex A. C., Pettibone G. W., Keay K., McManus P., Rex M. A., Ebersole J., Gallagher E. Distribution of indicator bacteria and Vibrio parahaemolyticus in sewage-polluted intertidal sediments. Appl Environ Microbiol. 1987 Aug;53(8):1756–1761. doi: 10.1128/aem.53.8.1756-1761.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K. A., Doyle J. D., King R. J., Seidler R. J., Stotzky G., Olsen R. H. Effects of 2,4-dichlorophenol, a metabolite of a genetically engineered bacterium, and 2,4-dichlorophenoxyacetate on some microorganism-mediated ecological processes in soil. Appl Environ Microbiol. 1991 Feb;57(2):412–418. doi: 10.1128/aem.57.2.412-418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain J. C., Pritchard P. H., Bourquin A. W. Effects of adaptation on biodegradation rates in sediment/water cores from estuarine and freshwater environments. Appl Environ Microbiol. 1980 Oct;40(4):726–734. doi: 10.1128/aem.40.4.726-734.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotzky G., Babich H. Survival of, and genetic transfer by, genetically engineered bacteria in natural environments. Adv Appl Microbiol. 1986;31:93–138. doi: 10.1016/s0065-2164(08)70440-4. [DOI] [PubMed] [Google Scholar]


