Abstract
Major histocompatibility complex (MHC) class II molecules bind antigenic peptides for display to T lymphocytes. Although the enzymes involved remain to be identified, it is commonly believed that class II associated peptides are released from intact antigens through a series of proteolytic steps carried out inside antigen presenting cells. We have examined the effect of amino acid substitutions on proteolytic processing of the model antigen hen-egg lysozyme (HEL). Altered HEL molecules, engineered by site-directed mutagenesis of a HEL cDNA, were expressed as separate stable transfectants in a B cell lymphoma line. Each transfectant processed a different mutant HEL protein for presentation on MHC class II. We purified the resulting class II-associated peptides and analyzed them by mass spectrometry. Our results strongly support the hypothesis that antigen processing continues after peptide binding to the MHC class II molecule and are most consistent with a scenario in which long peptides first bind to MHC class II and are then trimmed by exopeptidase.
It has been proposed that major histocompatibility complex (MHC) class II binding protects peptides from degradation during intracellular processing (1). This idea has been supported by the ability of affinity-purified class II molecules to protect bound peptides from protease digestion in vitro (2, 3). However, it is still unclear whether this protection occurs in vivo.
When bound, only the core of a peptide (about 9–10 amino acids) is in contact with the class II molecule, leaving the peptide ends free to extend beyond the binding site (4). Two lines of evidence suggest that these ends are available for trimming by exopeptidase enzymes. First, naturally processed peptides vary greatly in length, with most class II-associated peptides reported to span between 13 and 17 amino acids (5–7). Often naturally processed peptides belong to nested sets that share a common core sequence but differ at both the amino and carboxy termini. This length heterogeneity around a common core sequence suggests trimming of the peptide ends by exopeptidases.
Second, pool sequencing has revealed that proline residues occur with high frequency near the ends of naturally processed peptides, with proline especially common as the penultimate amino-terminal residue (8, 9). Proline is known to block the ability of several exopeptidase enzymes to remove amino- and carboxyl-terminal amino acids from peptides (10, 11) (due to the unusual chemical structure of the peptide bond formed by the imino group). The high frequency of proline residues near the ends of naturally processed peptides is consistent with the idea that these ends result from trimming by exopeptidase enzymes.
In this study, we examine the sequence specificity of antigen processing to obtain direct evidence for the amino-terminal trimming of naturally processed peptides.
MATERIALS AND METHODS
Expression Vector Assembly.
To create a membrane hen-egg lysozyme (mHEL) fusion construct, the entire gene for HEL was joined in-frame to a portion of the MHC class I Ld gene (see Fig. 1). A 4.0-kb KpnI fragment from the plasmid pLd-4 (12) containing part of the Ld genomic sequence was moved to Bluescript KS(−). A second NaeI site was created on this plasmid in the Ld α3 domain by oligonucleotide-directed site mutagenesis [changing nucleotides 2931–2933 as numbered by Linsk et al. (13)]. The resulting 3-kb NaeI fragment containing the 3′ end of the Ld gene was purified and inserted at the naturally occurring NaeI site found in the penultimate amino acid codon of a HEL cDNA, also in Bluescript KS(−). The codons for the final 2 amino acids of the HEL protein, lost after cleavage with NaeI, are restored in the fusion gene. Further site-directed mutagenesis was performed on this plasmid to generate a set of different mutant mHEL genes. For the final step in the construction, NotI–XhoI fragments containing the different HEL/Ld fusion genes were moved into the NotI–SalI sites of the expression vector pCEP4.
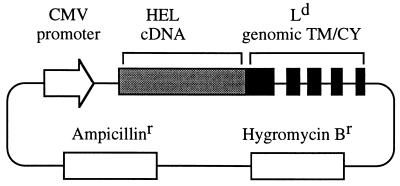
Figure 1.
Schematic representation of the mHEL expression vector. A cDNA encoding the entire HEL sequence (129 amino acids) was joined in-frame to a genomic portion of the MHC class I H-2Ld gene encoding a connecting stalk (8 amino acids), a transmembrane region (TM; 39 amino acids), and a complete cytoplasmic domain (CY; 25 amino acids). Transcription of the fusion gene was driven by the promoter region of the human cytomegalovirus (CMV) major immediate early gene. The construct extends through the genomic termination of transcription and polyadenylylation sequences used by Ld.
Transfection.
M12.C3.F6 cells (14) were transfected by electroporation and stable transfectants selected for hygromycin B resistance. For each transfection, 30 μg expression construct DNA in TE, 40 μg salmon-sperm carrier DNA, and 10 μl buffer (200 mM Hepes/1000 mM NaCl/50 mM KCl) were mixed in a total volume of 100 μl. The DNA mixture was added to 300 μl of DMEM supplemented with 5% fetal calf serum containing 1 × 107 M12.C3.F6 cells in a 2-mm gap disposable cuvette. After chilling on ice for 5 min, the cuvettes were electroporated at 200 V, 600 μF, in parallel with a 129-Ω resistance using an Electro Cell Manipulator 600 (BTX, San Diego). An average pulse lasted 8 msec. The cells were resuspended in 50 ml DMEM with 5% fetal calf serum and placed at 37C in a 5% CO2 incubator. After 24 h the cells were collected and placed in new medium containing 250 μg/ml of hygromycin B. Transfectants were identified by surface staining with a polyclonal rabbit antisera reactive to native HEL. All transfectants were able to process and present HEL 52–61 containing peptides to a standard T cell hybridoma (3A9), as described below.
Isolation and Characterization of Processed Peptides.
The I-Ak molecules were isolated from each transfectant (≈1 × 1010 cells, 40 liters) by affinity-chromatography using the mAb 40F (15) and the peptides extracted as described (16). The recovered peptides were first fractionated on reverse phase HPLC. Next, a T hybridoma bioassay was performed using 5 μl of each 250 μl HPLC fraction. The purified material was added to 1 × 105 3A9 cells (17) in a single well of a 96-well plate containing 200 μl of DMEM growth medium and 5 × 104 M12.C3.F6 cells as antigen-presenting cells (APC). After 24 h, the culture medium was harvested. The amount of interleukin 2 produced was measured by testing the supernatant’s ability to support the proliferation of an interleukin 2-dependent indicator line, CTLL. Recognition by 3A9 requires the HEL peptide to contain all residues from Asp-52 at the amino terminus through Arg-61 at the carboxy terminus (18). The 3A9 hybridoma responds about 30-fold better to the naturally processed HEL fragments, 48–62 and 48–63, than to the 52–61 core peptide due principally to the 30-fold higher binding strength of these peptides (19).
Analysis by Mass Spectrometry.
Each HPLC fraction that gave a positive response in the 3A9 T hybridoma assay was analyzed by matrix-assisted laser desorption ionization (MALDI)–time of flight mass spectrometry as follows; a portion (100 μl) was dried in a speed vac concentrator and resuspended in 8 μl of a 50% acetonitrile and 0.1% trifluoroacetic acid solution, 0.5 μl of this 8 μl was mixed with 0.5 μl of MALDI-Quality Alpha-CHC-Matrix Solution (Hewlett–Packard) on the sample plate of a Voyager RP (PerSeptive Diagnostics, Framingham, MA) mass spectrometer. Linear mass spectra were used for initial screening (acceleration potential 30 kV, ±m/z 1.0 accuracy), and reflector mass spectra (15 KV) were used for high accuracy (±m/z 0.2) molecular weight confirmation. For each transfectant, a single mixture was obtained by pooling an equal portion from each of the HEL 52–61 containing HPLC fractions. The relative yield of each peptide in this mixture was determined by integration of the peak areas in the linear mass spectrum of the mixture.
The following synthetic peptides were used to test the validity of our integration procedure; NPDGSTDYGILQINSR, TPGSTDYGILQINSR, NTDGSTDYGILQINSRW, and TDGSTDYGILQINSRW. The concentration of each peptide in solution was checked by comparison against a solution made using a control peptide, DGSSDYGILNINSRW. To do this, each peptide solution was mixed with an equal amount of the control peptide solution, and the ratio of the peak areas of the two peptides was determined from the mass spectrum. Next, the four test peptides were mixed in several different ratios (1:1:1:1, 2:2:1:1, 1:1:2:2, and others), and the ratio of the four peak areas in the mass spectra was determined. The peak areas were found to vary as predicted from the ratios of the dilutions. Therefore, we concluded that the ratios of the peak areas could be used to determine the relative peptide concentrations. In the following study, we identify a total of 24 HEL peptides, all of which are similar in sequence to the four test peptides. The shortest peptide spanned residues STDYGILQINSRW(W). The remaining peptides represent additions to this core sequence, with at most six additional residues at the amino terminus, the longest being NPNTDGSTDYGILQINSRW(W).
RESULTS
To explore the rules guiding amino-terminal proteolysis of peptides we examined the amino termini of peptides produced from a set of closely related HEL molecules engineered by site-directed mutagenesis. mHEL gene constructs (Fig. 1) were expressed in the I-Ak MHC class II bearing B-lymphoma line, M12.C3.F6. mHEL constructs were selected over secreted HEL constructs because they offered several advantages: fluorescence-activated cell sorting allowed easy isolation of the transfectants; an estimation of the expression level of the mHEL protein could be obtained from the antibody staining level; and the membrane tether prevented loss of the HEL protein into the medium. As demonstrated by flow cytometry, each transfectant expressed surface mHEL recognizable with a polyclonal rabbit antisera reactive to native HEL, and also with a conformationally dependent anti-HEL mAb, F10.6.6 (20) (data not shown). We found the endogenously synthesized mHEL to be efficiently processed for presentation on MHC class II, in agreement with previous work by Brooks et al. (21) performed with a similar mHEL construct.
Peptide-loaded I-Ak molecules were purified from each mHEL transfectant by affinity chromatography, and the peptides were released by mild acid denaturation. We focused on peptides containing HEL residues 52–61. Only these 10 amino acids residues, 52–61, are required for binding of this HEL region to I-Ak, with the aspartic acid side chain at position 52 (Asp-52) contributing most of the binding strength (22). To avoid interfering with the peptide’s ability to bind, all the mutations used in this study were confined to the peptide’s amino terminus (positions 42–48).
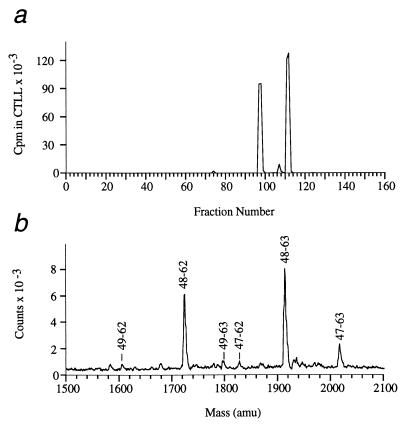
First, the recovered peptide mixtures were separated on HPLC, and then the fractions containing HEL 52–61 were identified using a T cell hybridoma bioassay. For example, the wild-type mHEL transfectant yielded four HPLC fractions that contained HEL 52–61 derived peptides (Fig. 2a). Mass spectrometry was used to identify the HEL peptides in these fractions by their unique molecular weights. In this case, the four HPLC fractions yielded six prominent HEL peptides. A mass spectrum containing these six peptides, obtained by mixing all four positive HPLC fractions, is shown in Fig. 2b. The relative amount of each HEL peptide was determined by integration of the peaks in the mass spectrum of this mixture (Table 1). For the wild-type mHEL transfectant, the two most prominent fragments (88%) started with Asp-48 and extended through residues Trp-62 or Trp-63 (i.e., DGSTDYGILQINSRW/W). It is important to note that these fragments, HEL 48–62 and 48–63, are identical to the two most prominent fragments previously recovered from B lymphoma cells cultured in exogenous lysozyme (16). Two minor versions of this epitope were also recovered, 9% beginning at Thr-47 (one residue longer), and 3% beginning at Gly-49 (one residue shorter). Other HEL 52–61 core peptide containing fragments were detected, but at much lower abundance; altogether these additional fragments represent less than 1% of the total.
Figure 2.
Identification of the HEL fragments presented on I-Ak by the wild-type mHEL transfectant. Peptides purified from the stable transfectant were separated into 160 fractions on reverse-phase HPLC as described. (a) Each fraction was tested for the ability to stimulate interleukin 2 production by the T cell hybridoma 3A9 (17), which is I-Ak restricted and recognizes residues 52–61 of HEL (18). In this experiment, four HPLC fractions contained HEL fragments; fractions 97, 98, 111, and 112. (b) Mass spectrum of a sample containing the six HEL peptides presented by the wild-type mHEL transfectant. This sample was obtained by mixing an equal amount from each of the four positive HPLC fractions identified in a. Each HEL fragment is labeled by its position within the HEL sequence. For example, residues 48–63 correspond to sequence DGSTDYGILQINSRWW. The relative amount of each HEL peptide was determined by integration of these peak areas (summarized in Table 1).
Table 1.
HEL peptides presented by the wild-type mHEL transfectant
| Relative yield, % | Position | Sequence
|
Mass, amu
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | Predicted | Observed | |||
| 9 | 2 | 47–62 | T | D | G | S | T | D | Y | G | I | L | Q | I | N | S | R | W | 1825.95 | 1825.66 | |
| 7 | 47–63 | T | D | G | S | T | D | Y | G | I | L | Q | I | N | S | R | W | W | 2011.17 | 2011.92 | |
| 88 | 32 | 48–62 | D | G | S | T | D | Y | G | I | L | Q | I | N | S | R | W | 1724.85 | 1724.86 | ||
| 56 | 48–63 | D | G | S | T | D | Y | G | I | L | Q | I | N | S | R | W | W | 1911.06 | 1910.86 | ||
| 3 | 1 | 49–62 | G | S | T | D | Y | G | I | L | Q | I | N | S | R | W | 1609.76 | 1609.84 | |||
| 2 | 49–63 | G | S | T | D | Y | G | I | L | Q | I | N | S | R | W | W | 1795.97 | 1795.85 | |||
Sequences of the six recovered HEL peptides are given in single-letter amino acid code. The numbers above each amino acid refer to its position within the HEL molecule. The predicted mass was calculated as the average isotopic mass of each peptide plus an additional hydrogen for ionization. The observed masses were obtained using matrix-assisted laser desorption ionization–time of flight as described. The relative yield for each peptide was calculated by integration of the mass spectrum shown in Fig. 2b.
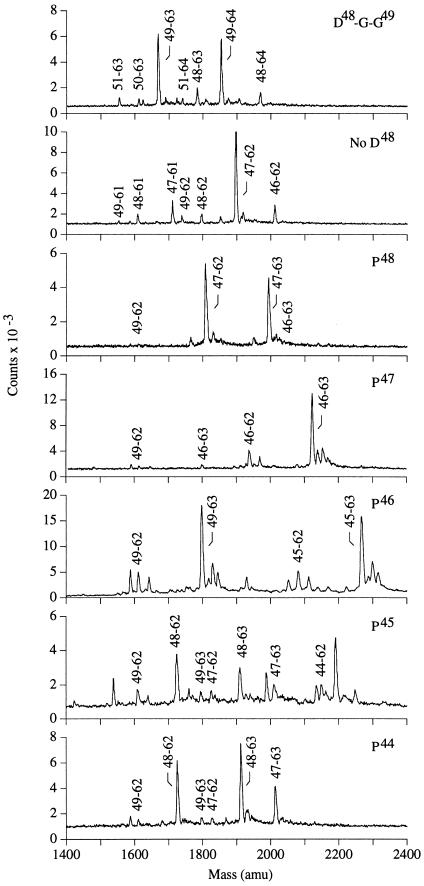
Two mHEL mutants were made to test whether the cleavage observed between residues Thr-47 and Asp-48 was sequence-specific. In one, an additional glycine was inserted between Asp-48 and Gly-49 and in another, residue Asp-48 was deleted. The main cleavage for these mutants, like the wild-type, occurred four amino acids before the P1 anchor residue at Asp-52 (see D48-G-G49 and No D48 in Fig. 3, summarized in Table 2). Because these cleavages were not directed by the sequence of the bound peptide, we conclude that the 4 amino acids between the amino terminus and the P1 anchor represent the extent of protection afforded by the peptide binding groove of the MHC class II molecule. The minor start sites probably result from the inability of the MHC class II molecule to completely protect the peptide from proteolytic activity.
Figure 3.
Mass spectrum of HEL 52–61 containing peptides recovered from the mHEL mutants. Each panel corresponds to a different mHEL mutant. For each mutant, the sample was obtained by mixing an equal amount from all the HPLC fractions able to stimulate the HEL 52–61 reactive T cell hybridoma, 3A9. The observed mass and relative yield of each HEL fragment is given in Table 2.
Table 2.
HEL peptides presented by the mutant mHEL transfectants
| mHEL Mutant | Relative yield, % | Position | Sequence | Mass, amu
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted | Observed | |||||||||||||||||
| 48 | 49 | 50 | 51 | 52 | 53 | — | 62 | 63 | 64 | |||||||||
| D48-G-G49 | 14 | 8 | 48–63 | D | G | G | S | T | D | — | R | W | 1781.90 | 1781.84 | ||||
| 6 | 48–64 | D | G | G | S | T | D | — | R | W | W | 1968.11 | 1968.11 | |||||
| 79 | 41 | 49–63 | G | G | S | T | D | — | R | W | 1666.81 | 1666.85 | ||||||
| 38 | 49–64 | G | G | S | T | D | — | R | W | W | 1853.02 | 1852.99 | ||||||
| 3 | 2 | 50–63 | G | S | T | D | — | R | W | 1609.76 | 1609.95 | |||||||
| 1 | 50–64 | G | S | T | D | — | R | W | W | 1795.97 | 1796.16 | |||||||
| 5 | 3 | 51–63 | S | T | D | — | R | W | 1552.70 | 1552.89 | ||||||||
| 2 | 51–64 | S | T | D | — | R | W | W | 1738.92 | 1738.85 | ||||||||
| 46 | 47 | 48 | 49 | 50 | 51 | — | 60 | 61 | 62 | |||||||||
| No D48 | 12 | 1 | 46–61 | N | T | G | S | T | D | — | R | W | 1824.97 | 1826.58 | ||||
| 11 | 46–62 | N | T | G | S | T | D | — | R | W | W | 2011.18 | 2011.41 | |||||
| 72 | 11 | 47–61 | T | G | S | T | D | — | R | W | 1710.86 | 1710.79 | ||||||
| 61 | 47–62 | T | G | S | T | D | — | R | W | W | 1897.08 | 1896.89 | ||||||
| 10 | 6 | 48–61 | G | S | T | D | — | R | W | 1609.76 | 1609.73 | |||||||
| 4 | 48–62 | G | S | T | D | — | R | W | W | 1795.97 | 1795.81 | |||||||
| 5 | 2 | 49–61 | S | T | D | — | R | W | 1552.70 | 1553.24 | ||||||||
| 4 | 49–62 | S | T | D | — | R | W | W | 1738.92 | 1738.73 | ||||||||
| 46 | 47 | 48 | 49 | 50 | 51 | 52 | — | 61 | 62 | 63 | ||||||||
| P48 | 0.5 | 0 | 46–62 | N | T | P | G | S | T | D | — | R | W | 1922.06 | — | |||
| 0.5 | 46–63 | N | T | P | G | S | T | D | — | R | W | W | 2108.30 | 2109.81 | ||||
| 99 | 54 | 47–62 | T | P | G | S | T | D | — | R | W | 1807.98 | 1808.13 | |||||
| 45 | 47–63 | T | P | G | S | T | D | — | R | W | W | 1994.19 | 1994.33 | |||||
| 0 | 0 | 48–62 | P | G | S | T | D | — | R | W | 1706.88 | — | ||||||
| 0 | 48–63 | P | G | S | T | D | — | R | W | W | 1893.09 | — | ||||||
| 0.5 | 0.5 | 49–62 | G | S | T | D | — | R | W | 1609.76 | 1612.23 | |||||||
| 0 | 49–63 | G | S | T | D | — | R | W | W | 1795.97 | — | |||||||
| 46 | 47 | 48 | 49 | 50 | 51 | 52 | — | 61 | 62 | 63 | ||||||||
| P47 | 96 | 19 | 46–62 | N | P | D | G | S | T | D | — | R | W | 1936.07 | 1937.26 | |||
| 73 | 46–63 | N | P | D | G | S | T | D | — | R | W | W | 2122.28 | 2121.86 | ||||
| 0 | 0 | 47–62 | P | D | G | S | T | D | — | R | W | 1821.96 | — | |||||
| 0 | 47–63 | P | D | G | S | T | D | — | R | W | W | 2008.18 | — | |||||
| 0 | 0 | 48–62 | D | G | S | T | D | — | R | W | 1724.85 | — | ||||||
| 0 | 48–63 | D | G | S | T | D | — | R | W | W | 1911.06 | — | ||||||
| 4 | 2 | 49–62 | G | S | T | D | — | R | W | 1609.76 | 1610.03 | |||||||
| 2 | 49–63 | G | S | T | D | — | R | W | W | 1795.97 | 1796.61 | |||||||
| 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | — | 61 | 62 | 63 | |||||||
| P46 | 47 | 9 | 45–62 | R | P | T | D | G | S | T | D | — | R | W | 2079.26 | 2080.77 | ||
| 38 | 45–63 | R | P | T | D | G | S | T | D | — | R | W | W | 2265.47 | 2267.37 | |||
| 0 | 0 | 46–62 | P | T | D | G | S | T | D | — | R | W | 1923.10 | — | ||||
| 0 | 46–63 | P | T | D | G | S | T | D | — | R | W | W | 2109.28 | — | ||||
| 0 | 0 | 47–62 | T | D | G | S | T | D | — | R | W | 1825.95 | — | |||||
| 0 | 47–63 | T | D | G | S | T | D | — | R | W | W | 2012.17 | — | |||||
| 1 | 0 | 48–62 | D | G | S | T | D | — | R | W | 1706.88 | — | ||||||
| 1 | 48–63 | D | G | S | T | D | — | R | W | W | 1893.09 | 1893.86 | ||||||
| 53 | 9 | 49–62 | G | S | T | D | — | R | W | 1609.76 | 1610.81 | |||||||
| 44 | 49–63 | G | S | T | D | — | R | W | W | 1795.97 | 1797.21 | |||||||
| 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | — | 61 | 62 | 63 | ||||||
| P45 | 11 | 11 | 44–62 | N | P | N | T | D | G | S | T | D | — | R | W | 2151.28 | 2151.80 | |
| 0 | 44–63 | N | P | N | T | D | G | S | T | D | — | R | W | W | 2337.49 | — | ||
| 0 | 0 | 45–62 | P | N | T | D | G | S | T | D | — | R | W | 2037.17 | — | |||
| 0 | 45–63 | P | N | T | D | G | S | T | D | — | R | W | W | 2223.39 | — | |||
| 0 | 0 | 46–62 | N | T | D | G | S | T | D | — | R | W | 1940.06 | — | ||||
| 0 | 46–63 | N | T | D | G | S | T | D | — | R | W | W | 2126.27 | — | ||||
| 21 | 8 | 47–62 | T | D | G | S | T | D | — | R | W | 1825.95 | 1826.52 | |||||
| 13 | 47–63 | T | D | G | S | T | D | — | R | W | W | 2012.17 | 2011.96 | |||||
| 55 | 31 | 48–62 | D | G | S | T | D | — | R | W | 1724.85 | 1724.65 | ||||||
| 24 | 48–63 | D | G | S | T | D | — | R | W | W | 1911.06 | 1910.64 | ||||||
| 12 | 6 | 49–62 | G | S | T | D | — | R | W | 1609.76 | 1609.59 | |||||||
| 6 | 49–63 | G | S | T | D | — | R | W | W | 1795.97 | 1795.68 | |||||||
| 47 | 48 | 49 | 50 | 51 | 52 | — | 61 | 62 | 63 | |||||||||
| P44 | 19 | 2 | 47–62 | T | D | G | S | T | D | — | R | W | 1825.95 | 1825.70 | ||||
| 17 | 47–63 | T | D | G | S | T | D | — | R | W | W | 2012.17 | 2011.30 | |||||
| 77 | 34 | 48–62 | D | G | S | T | D | — | R | W | 1724.85 | 1724.58 | ||||||
| 43 | 48–63 | G | S | T | D | — | R | W | W | 1911.06 | 1910.47 | |||||||
| 4 | 1 | 49–62 | G | S | T | D | — | R | W | 1609.76 | 1609.84 | |||||||
| 3 | 49–63 | G | S | T | D | — | R | W | W | 1795.97 | 1796.37 | |||||||
Sequences of the 6 recovered HEL peptides given in single letter amino acid code. (—), a fragment that was not recovered. Amino acids changed by mutagenesis given in bold.
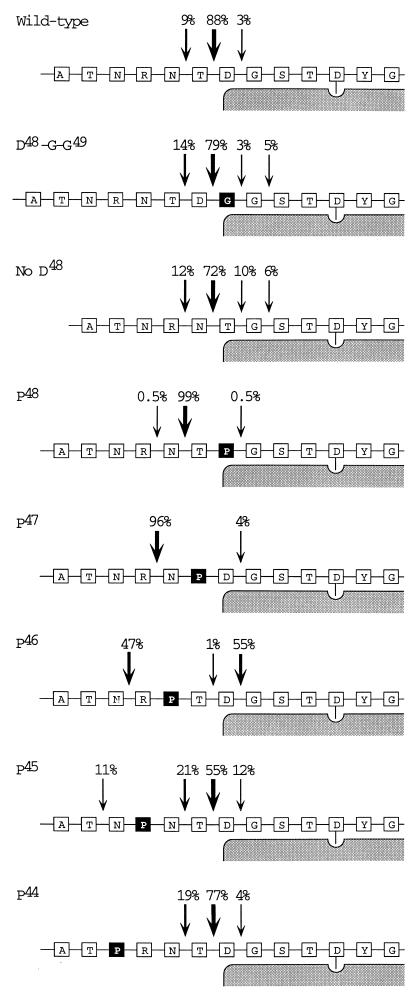
Additional mHEL transfectants were made with proline substitutions near the amino terminus. Proline is known to inhibit the activity of a number of aminopeptidase enzymes, and we sought to test whether proline could block the aminopeptidase activity observed above. Proline substitution at position 48 caused essentially all of the presented peptides to be one residue longer than wild type (99%, TPGST… , see P48 in Fig. 3, summarized in Table 2). Similarly, almost all of the peptides presented by the Pro-47 transfectant were two residues longer (96%, NPDGST… ). We interpret these results to indicate that proline can act as a stop signal for aminopeptidase trimming in this system.
The ability of proline to protect the amino terminus decreased as the distance from the MHC class II molecule increased. Only 47% of the fragments produced from the Pro-46 mutant contained the proline (RPTDGST… ), 11% from the Pro-45 mutant (NPNTDGST… ), and none from the Pro-44 mutant (summarized in Fig. 4). This loss of protection implies the involvement of a second peptidase, either an endopeptidase or possibly a dipeptidyl peptidase, able to remove proline residues that lie sufficiently far from the class II molecule.
Figure 4.
Summary of HEL peptides presented by the different mHEL transfectants. Each series of boxes represents a different mHEL mutant, with the amino acid sequence given in single letter code. The shaded region represents the MHC class II molecule. The aspartic acid anchor at P1 (Asp-52) is shown with its side chain extending into the class II molecule. The amino acids altered by site-directed mutagenesis are shown as dark boxes. Arrows point to the amino termini of the recovered peptides. Percentages, given above each arrow, were obtained by integration of mass spectra as described in Fig. 2b and Table 1.
It is not possible to explain these results as a change in peptide binding strength caused by the proline substitutions. Peptides corresponding to the major HEL fragments recovered from the Pro-46 mutant (RPTDGST—), Pro-47 mutant (NPDGST—), and Pro-48 mutant (TPGST—) were synthesized. To avoid problems with solubility, these peptides were made with carboxy termini at Arg-61 rather than Trp-62. Each was tested for the ability to inhibit the binding of a radiolabeled standard peptide (YEDYGILQINSR) to I-Ak. No differences were observed between the ability of these peptides, and the wild-type 48–61 peptide, to compete for I-Ak binding (data not shown).
We previously observed that most naturally processed I-Ak associated peptides contain 3–4 amino acids between the amino terminus and the P1 anchor residue (22). The only exceptions, peptides with penultimate prolines at the amino terminus, have all been 1–2 amino acids longer. The inability of proline to protect longer peptides from amino-terminal trimming fits well with this previous observation.
DISCUSSION
Taken together, these results highlight the role MHC class II molecules play in protecting immunogenic peptides from complete proteolysis during antigen processing. Although additional work will be needed to identify the enzymes involved and their subcellular sites of action, the ability to detect small changes in naturally processed peptides should help to clarify the mechanisms by which APC cleave, bind, and trim antigen fragments for presentation on MHC class II molecules.
We envision the following scenario for the antigen processing of HEL. After internalization by the APC, a combination of disulfide bond reduction and low pH denaturation lead to an unfolding of the native HEL protein. This unfolding allows the MHC class II molecule to bind large HEL fragments, or possibly the entire HEL protein. Ample evidence exists for the binding of unfolded HEL to I-Ak (for example, see ref. 23). Once bound, the HEL segment is protected from catabolism; the extent of protection dictated by the dimensions of the peptide-combining site of I-Ak molecule.
An alternative explanation for these results would be that fragments are cut and trimmed before binding. Once bound to the class II molecule, these fragments would be transported to the cell surface without further change. We believe the length of fragments (4 amino acids between the amino terminus and the P1 anchor residue), as well as the limited protection afforded by the proline substitutions (at most, an additional 4 amino acids) argues against this interpretation.
Our working hypothesis is that the majority of peptide trimming takes place in an intracellular compartment, and further studies to address this issue are underway. A recent study based on a functional T cell readout has suggested that peptides bound to MHC class II can be trimmed at the cell surface (24). However, in preliminary experiments we have been unable to demonstrate cell surface trimming. In these experiments, radiolabeled peptides were first offered to APC and then recovered by immunoprecipitation of the MHC class II molecules for sequencing by Edman degradation.
To our knowledge, this is the first reported characterization of the naturally processed peptides obtained using a set of engineered antigens. Previously, the biochemical analysis of naturally processed peptides was limited to the few proteins (16, 25) that could be obtained in large enough quantities to allow sufficient loading of MHC class II by the exogenous pathway. Our current approach avoids this limitation by using the APC to synthesize the antigen. We believe it should be possible to apply this procedure to any antigen that can correctly fold and traffic to the cell surface.
Acknowledgments
We thank Anthony Combs for preparing the synthetic peptides used in this study, Dr. T. H. Hansen for his gift of the pLd4 plasmid, and Janet Casmaer for her help in preparing the manuscript.
Footnotes
.
Abbreviations: HEL, hen-egg lysozyme; mHEL, membrane HEL; MHC, major histocompatibility complex; APC, antigen-presenting cells.
References
- 1.Werdelin O. Scand J Immunol. 1986;24:625–636. doi: 10.1111/j.1365-3083.1986.tb02181.x. [DOI] [PubMed] [Google Scholar]
- 2.Donermeyer D L, Allen P M. J Immunol. 1989;142:1063–1068. [PubMed] [Google Scholar]
- 3.Mouritsen S, Meldal M, Werdelin O, Hansen A S, Buus S. J Immunol. 1992;149:1987–1993. [PubMed] [Google Scholar]
- 4.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 5.Rudensky A Y, Preston-Hurlburt P, Hong S C, Barlow A, Janeway C A., Jr Nature (London) 1991;353:622–627. doi: 10.1038/353622a0. [DOI] [PubMed] [Google Scholar]
- 6.Hunt D F, Michel H, Dickinson T A, Shabanowitz J, Cox A L, Sakaguchi K, Appella E, Grey H M, Sette A. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 7.Rammensee H G, Friede T, Stevanovic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 8.Kropshofer H, Max H, Halder T, Kalbus M, Muller C A, Kalbacher H. J Immunol. 1993;151:4732–4742. [PubMed] [Google Scholar]
- 9.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee H G. Immunogenetics. 1994;39:230–242. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]
- 10.Ansorge S, Schon E, Kunz D. Biomed Biochim Acta. 1991;50:799–807. [PubMed] [Google Scholar]
- 11.McDonald J K, Barrett A J, editors. Mammalian Proteases. Vol. 2. New York: Academic; 1986. pp. 23–100. [Google Scholar]
- 12.Evans G A, Margulies D H, Shykind B, Seidman J G, Ozato K. Nature (London) 1982;300:755–757. doi: 10.1038/300755a0. [DOI] [PubMed] [Google Scholar]
- 13.Linsk R, Vogel J, Stauss H, Forman J, Goodenow R S. J Exp Med. 1986;164:794–813. doi: 10.1084/jem.164.3.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wade W F, Chen Z Z, Maki R, McKercher S, Palmer E, Cambier J C, Freed J H. Proc Natl Acad Sci USA. 1989;86:6297–6301. doi: 10.1073/pnas.86.16.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierres M, Devaux C, Dosseto M, Marchetto S. Immunogenetics. 1981;14:481–495. doi: 10.1007/BF00350120. [DOI] [PubMed] [Google Scholar]
- 16.Nelson C A, Roof R W, McCourt D W, Unanue E R. Proc Natl Acad Sci USA. 1992;89:7380–7383. doi: 10.1073/pnas.89.16.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen P M, Unanue E R. J Immunol. 1984;132:1077–1079. [PubMed] [Google Scholar]
- 18.Allen P M, Matsueda G R, Evans R J, Dunbar J B, Jr, Marshall G R, Unanue E R. Nature (London) 1987;327:713–715. doi: 10.1038/327713a0. [DOI] [PubMed] [Google Scholar]
- 19.Nelson C A, Petzold S J, Unanue E R. Proc Natl Acad Sci USA. 1993;90:1227–1231. doi: 10.1073/pnas.90.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischmann T, Souchon H, Riottot M-M, Tello D, Poljak R J. J Mol Biol. 1988;203:527–529. doi: 10.1016/0022-2836(88)90022-8. [DOI] [PubMed] [Google Scholar]
- 21.Brooks A, Hartley S, Kjer-Nielsen L, Perera J, Goodnow C C, Basten A, McCluskey J. Proc Natl Acad Sci USA. 1991;88:3290–3294. doi: 10.1073/pnas.88.8.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson C A, Viner N J, Young S P, Petzold S J, Unanue E R. J Immunol. 1996;157:755–762. [PubMed] [Google Scholar]
- 23.Lindner, R. & Unanue, E. R. (1996) EMBO J., in press. [PMC free article] [PubMed]
- 24.Larsen S L, Pedersen L, O, Buss S, Stryhn A. J Exp Med. 1996;184:183–189. doi: 10.1084/jem.184.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore S E H, Schofield J P, Watts C. Immunology. 1995;85:523–530. [PMC free article] [PubMed] [Google Scholar]