Abstract
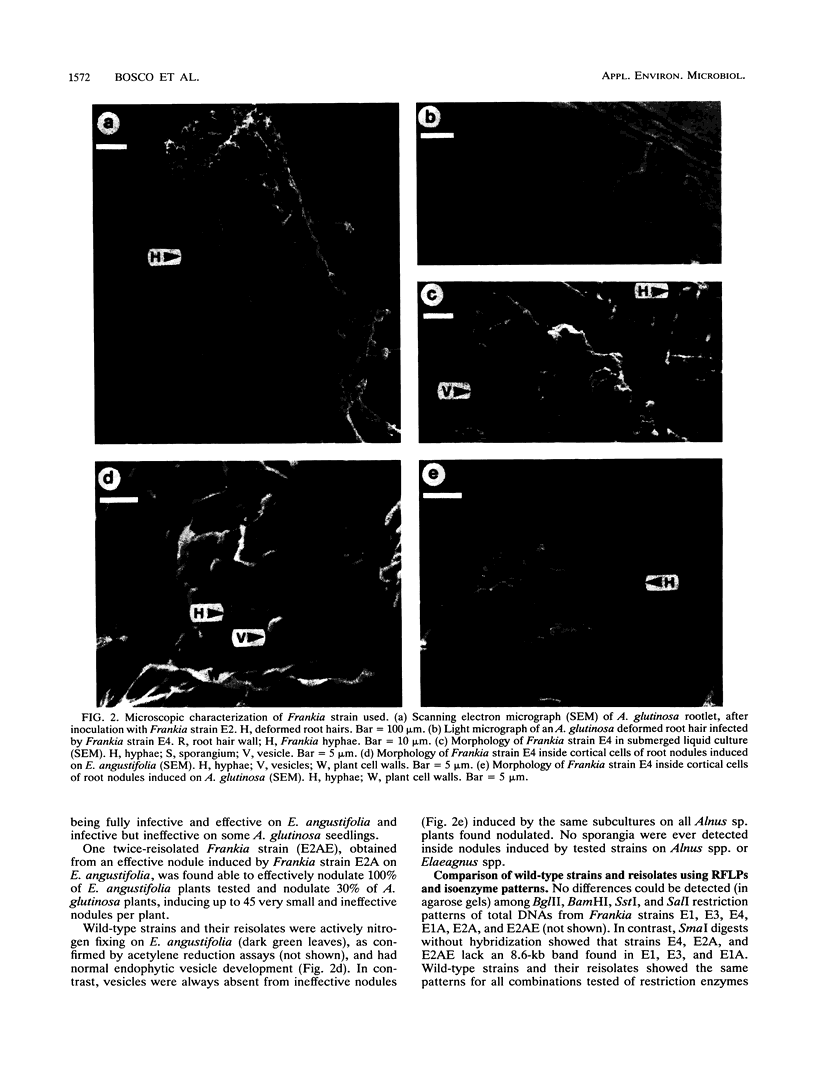
Phenotypic and genotypic methods were used to prove the existence of Frankia strains isolated from an Elaeagnus sp. that are able to cross the inoculation barriers and infect Alnus spp. also. Repeated cycles of inoculation, nodulation, and reisolation were performed under axenic conditions. Frankia wild-type strain UFI 13270257 and three of its coisolates did exhibit complete infectivity and effectiveness on Elaeagnus spp. and Hippophaë rhamnoides and variable infectivity on Alnus spp. Microscopical observation of host plant roots showed that these strains are able to infect Alnus spp. by penetrating deformed root hairs. Reisolates obtained from nodules induced on monoxenic Alnus glutinosa, Alnus incana, and Elaeagnus angustifolia resembled the parent strains in host infectivity range, in planta and in vitro morphophysiology, isoenzymes, and nif and rrn restriction fragment length polymorphisms, thus fulfilling Koch's postulates on both host plant genera. Alnus and Elaeagnus group-specific polymerase chain reaction DNA amplifications, DNA-DNA hybridizations, and partial gene sequences coding for 16S rRNA provided evidence for the genetic uniformity of wild-type strains and their inclusion into one and the same genomic species, clearly belonging to the Elaeagnus group of Frankia species.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloom R. A., Mullin B. C., Tate R. L., 3rd DNA restriction patterns and DNA-DNA solution hybridization studies of Frankia isolates from Myrica pennsylvanica (bayberry). Appl Environ Microbiol. 1989 Sep;55(9):2155–2160. doi: 10.1128/aem.55.9.2155-2160.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaham D., Deltredici P., Torrey J. G. Isolation and Cultivation in vitro of the Actinomycete Causing Root Nodulation in Comptonia. Science. 1978 Feb 24;199(4331):899–902. doi: 10.1126/science.199.4331.899. [DOI] [PubMed] [Google Scholar]
- Gardes M., Bousquet J., Lalonde M. Isozyme Variation among 40 Frankia Strains. Appl Environ Microbiol. 1987 Jul;53(7):1596–1603. doi: 10.1128/aem.53.7.1596-1603.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Nazaret S., Cournoyer B., Normand P., Simonet P. Phylogenetic relationships among Frankia genomic species determined by use of amplified 16S rDNA sequences. J Bacteriol. 1991 Jul;173(13):4072–4078. doi: 10.1128/jb.173.13.4072-4078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P., Cournoyer B., Simonet P., Nazaret S. Analysis of a ribosomal RNA operon in the actinomycete Frankia. Gene. 1992 Feb 1;111(1):119–124. doi: 10.1016/0378-1119(92)90612-s. [DOI] [PubMed] [Google Scholar]
- Normand P., Simonet P., Bardin R. Conservation of nif sequences in Frankia. Mol Gen Genet. 1988 Aug;213(2-3):238–246. doi: 10.1007/BF00339587. [DOI] [PubMed] [Google Scholar]
- Pernodet J. L., Boccard F., Alegre M. T., Blondelet-Rouault M. H., Guérineau M. Resistance to macrolides, lincosamides and streptogramin type B antibiotics due to a mutation in an rRNA operon of Streptomyces ambofaciens. EMBO J. 1988 Jan;7(1):277–282. doi: 10.1002/j.1460-2075.1988.tb02810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prin Y., Duhoux E., Diem H. G., Roederer Y., Dommergues Y. R. Aerial Nodules in Casuarina cunninghamiana. Appl Environ Microbiol. 1991 Mar;57(3):871–874. doi: 10.1128/aem.57.3.871-874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet P., Grosjean M. C., Misra A. K., Nazaret S., Cournoyer B., Normand P. Frankia genus-specific characterization by polymerase chain reaction. Appl Environ Microbiol. 1991 Nov;57(11):3278–3286. doi: 10.1128/aem.57.11.3278-3286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen D., Jones C., Schofield J. P. A rapid method for isolating high quality plasmid DNA suitable for DNA sequencing. Nucleic Acids Res. 1990 Dec 25;18(24):7463–7464. doi: 10.1093/nar/18.24.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983 Jul 1;132(1):14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]