Abstract
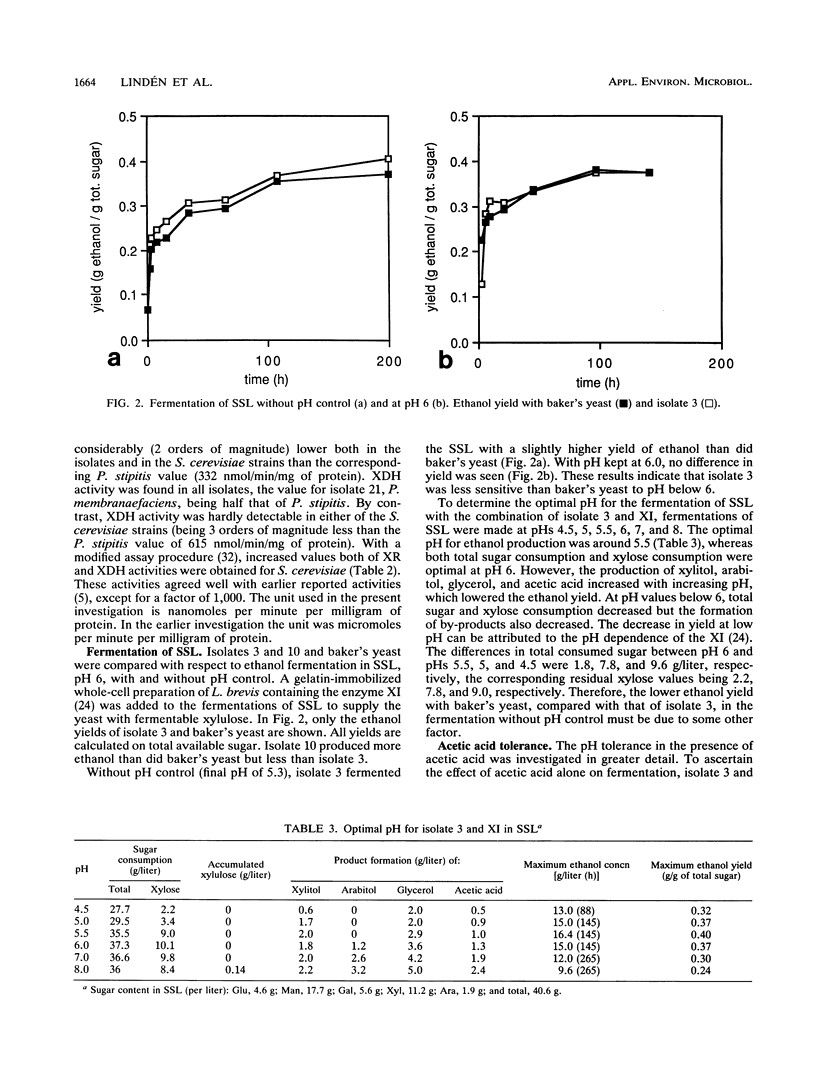
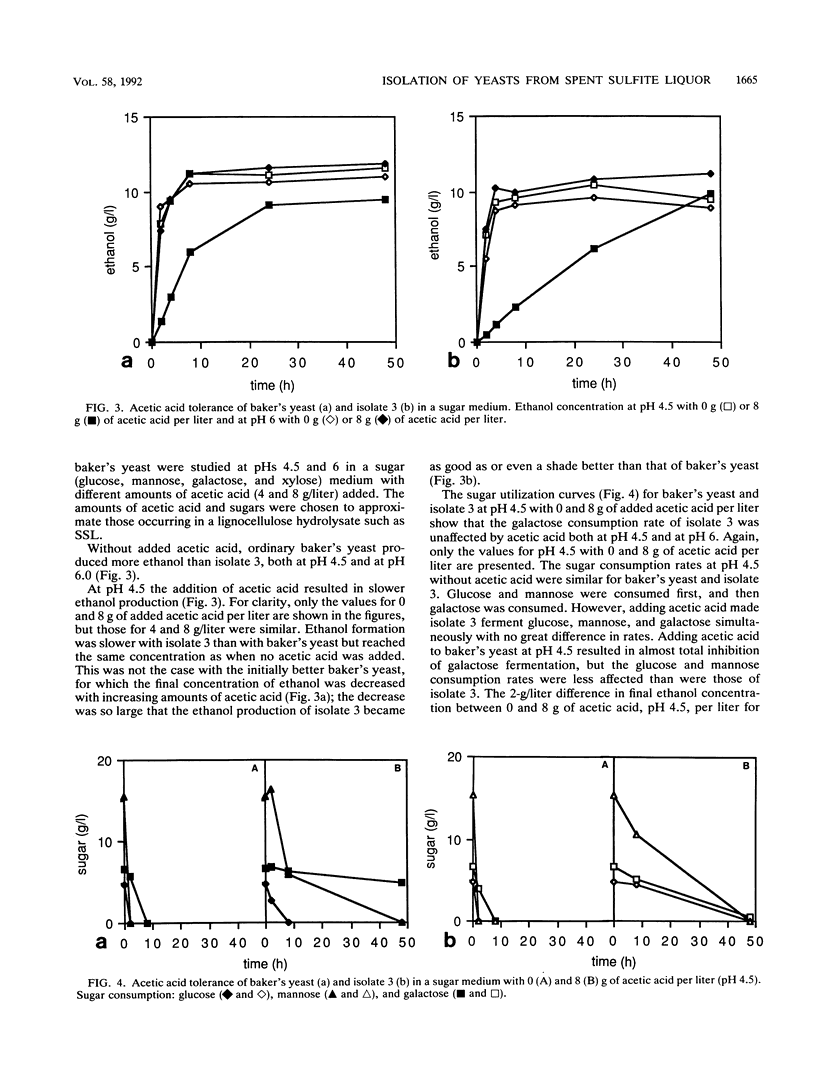
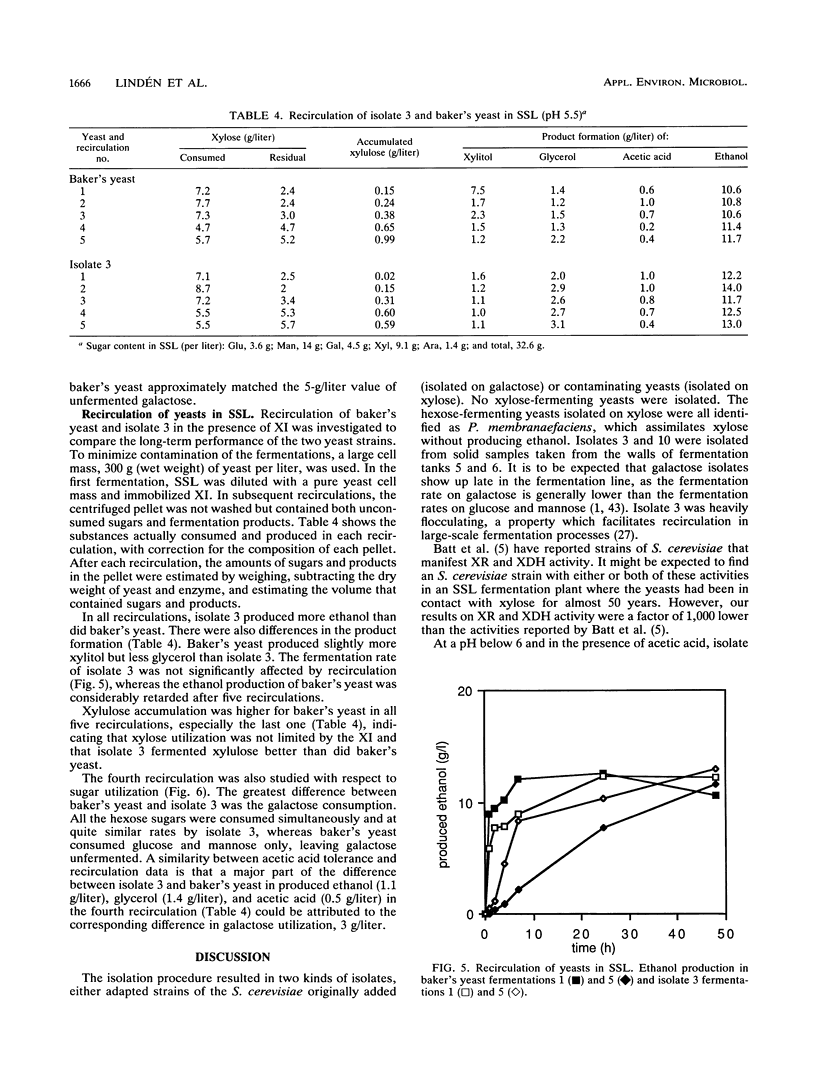
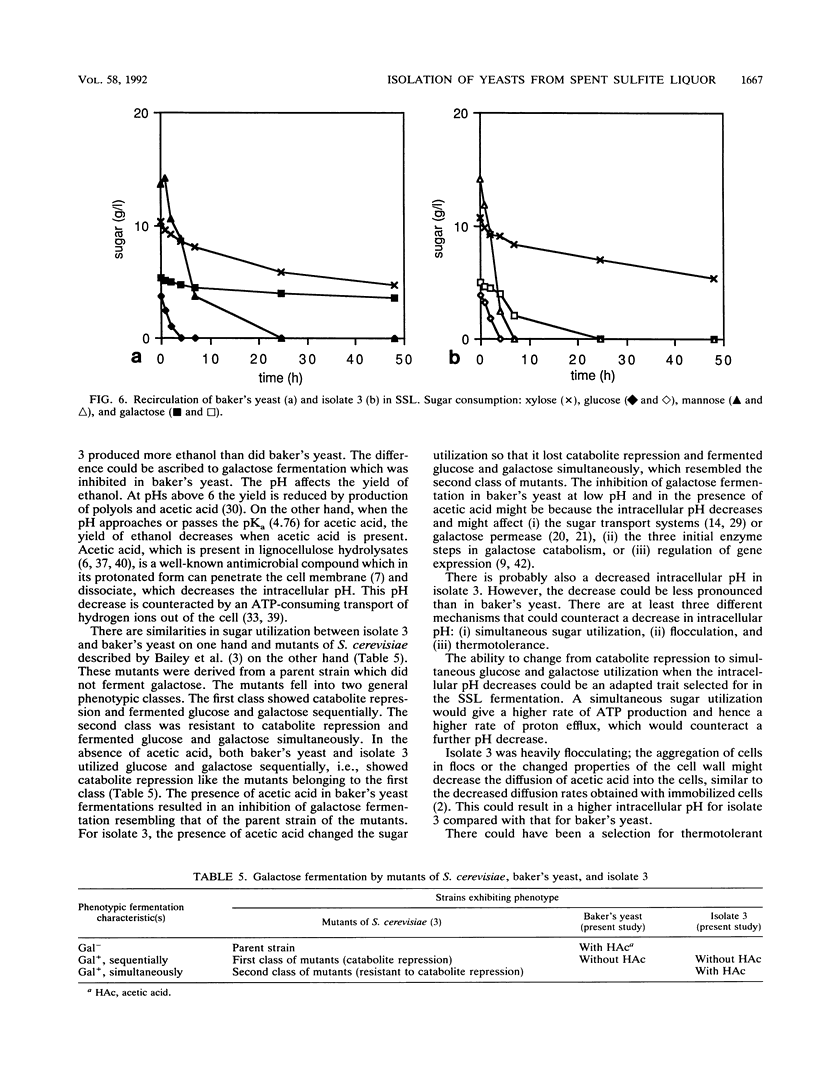
From a continuous spent sulfite liquor fermentation plant, two species of yeast were isolated, Saccharomyces cerevisiae and Pichia membranaefaciens. One of the isolates of S. cerevisiae, no. 3, was heavily flocculating and produced a higher ethanol yield from spent sulfite liquor than did commercial baker's yeast. The greatest difference between isolate 3 and baker's yeast was that of galactose fermentation, even when galactose utilization was induced, i.e., when they were grown in the presence of galactose, prior to fermentation. Without acetic acid present, both baker's yeast and isolate 3 fermented glucose and galactose sequentially. Galactose fermentation with baker's yeast was strongly inhibited by acetic acid at pH values below 6. Isolate 3 fermented galactose, glucose, and mannose without catabolite repression in the presence of acetic acid, even at pH 4.5. The xylose reductase (EC 1.1.1.21) and xylitol dehydrogenase (EC 1.1.1.9) activities were determined in some of the isolates as well as in two strains of S. cerevisiae (ATCC 24860 and baker's yeast) and Pichia stipitis CBS 6054. The S. cerevisiae strains manifested xylose reductase activity that was 2 orders of magnitude less than the corresponding P. stipitis value of 890 nmol/min/mg of protein. The xylose dehydrogenase activity was 1 order of magnitude less than the corresponding activity of P. stipitis (330 nmol/min/mg of protein).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey R. B., Benitez T., Woodward A. Saccharomyces cerevisiae Mutants Resistant to Catabolite Repression: Use in Cheese Whey Hydrolysate Fermentation. Appl Environ Microbiol. 1982 Sep;44(3):631–639. doi: 10.1128/aem.44.3.631-639.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth I. R. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985 Dec;49(4):359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carlson M. Regulation of sugar utilization in Saccharomyces species. J Bacteriol. 1987 Nov;169(11):4873–4877. doi: 10.1128/jb.169.11.4873-4877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote P. J., Cole M. B., Jones M. V. Induction of increased thermotolerance in Saccharomyces cerevisiae may be triggered by a mechanism involving intracellular pH. J Gen Microbiol. 1991 Jul;137(7):1701–1708. doi: 10.1099/00221287-137-7-1701. [DOI] [PubMed] [Google Scholar]
- Does A. L., Bisson L. F. Comparison of glucose uptake kinetics in different yeasts. J Bacteriol. 1989 Mar;171(3):1303–1308. doi: 10.1128/jb.171.3.1303-1308.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallborn J., Walfridsson M., Airaksinen U., Ojamo H., Hahn-Hägerdal B., Penttilä M., Keräsnen S. Xylitol production by recombinant Saccharomyces cerevisiae. Biotechnology (N Y) 1991 Nov;9(11):1090–1095. doi: 10.1038/nbt1191-1090. [DOI] [PubMed] [Google Scholar]
- Jeffries T. W. Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng Biotechnol. 1983;27:1–32. doi: 10.1007/BFb0009101. [DOI] [PubMed] [Google Scholar]
- Kou S. C., Christensen M. S., Cirillo V. P. Galactose transport in Saccharomyces cerevisiae. II. Characteristics of galactose uptake and exchange in galactokinaseless cells. J Bacteriol. 1970 Sep;103(3):671–678. doi: 10.1128/jb.103.3.671-678.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Cirillo V. P. Galactose transport in Saccharomyces cerevisiae. 3. Characteristics of galactose uptake in transferaseless cells: evidence against transport-associated phosphorylation. J Bacteriol. 1970 Sep;103(3):679–685. doi: 10.1128/jb.103.3.679-685.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötter P., Amore R., Hollenberg C. P., Ciriacy M. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet. 1990 Dec;18(6):493–500. doi: 10.1007/BF00327019. [DOI] [PubMed] [Google Scholar]
- Romano A. H. Facilitated diffusion of 6-deoxy-D-glucose in bakers' yeast: evidence against phosphorylation-associated transport of glucose. J Bacteriol. 1982 Dec;152(3):1295–1297. doi: 10.1128/jb.152.3.1295-1297.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog K., Hahn-Hägerdal B. Effect of Oxygenation on Xylose Fermentation by Pichia stipitis. Appl Environ Microbiol. 1990 Nov;56(11):3389–3394. doi: 10.1128/aem.56.11.3389-3394.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuma S., Nakashima N., Tantirungkij M., Kinoshita S., Okada H., Seki T., Yoshida T. Isolation of xylose reductase gene of Pichia stipitis and its expression in Saccharomyces cerevisiae. Appl Biochem Biotechnol. 1991 Spring;28-29:327–340. doi: 10.1007/BF02922612. [DOI] [PubMed] [Google Scholar]
- Warth A. D. Effect of benzoic acid on glycolytic metabolite levels and intracellular pH in Saccharomyces cerevisiae. Appl Environ Microbiol. 1991 Dec;57(12):3415–3417. doi: 10.1128/aem.57.12.3415-3417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills C. Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1990;25(4):245–280. doi: 10.3109/10409239009090611. [DOI] [PubMed] [Google Scholar]



