Abstract
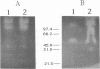
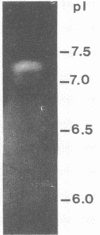
The involvement of Rhizobium enzymes that degrade plant cell wall polymers has long been an unresolved question about the infection process in root nodule symbiosis. Here we report the production of enzymes from Rhizobium leguminosarum bv. trifolii that degrade carboxymethyl cellulose and polypectate model substrates with sensitive methods that reliably detect the enzyme activities: a double-layer plate assay, quantitation of reducing sugars with a bicinchoninate reagent, and activity gel electrophoresis-isoelectric focusing. Both enzyme activities were (i) produced commonly by diverse wild-type strains, (ii) cell bound with at least some of the activity associated with the cell envelope, and (iii) not changed appreciably by growth in the presence of the model substrates or a flavone that activates expression of nodulation (nod) genes on the resident symbiotic plasmid (pSym). Equivalent levels of carboxymethyl cellulase activity were found in wild-type strain ANU843 and its pSym-cured derivative, ANU845, consistent with previous results of Morales et al. (V. Morales, E. Martínez-Molina, and D. Hubbell, Plant Soil 80:407-415, 1984). However, polygalacturonase activity was lower in ANU845 and was not restored to wild-type levels in the recombinant derivative of pSym- ANU845 containing the common and host-specific nod genes within a 14-kb HindIII DNA fragment of pSym from ANU843 cloned on plasmid pRt032. Activity gel electrophoresis resolved three carboxymethyl cellulase isozymes of approximately 102, 56, and 33 kDa in cell extracts from ANU843. Isoelectric focusing activity gels revealed one ANU843 polygalacturonase isozyme with a pI of approximately 7.2. These studies show that R. leguminosarum bv. trifolii produces multiple enzymes that cleave glycosidic bonds in plant cell walls and that are cell bound.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Dazzo F. B., Brill W. J. Bacterial polysaccharide which binds Rhizobium trifolii to clover root hairs. J Bacteriol. 1979 Mar;137(3):1362–1373. doi: 10.1128/jb.137.3.1362-1373.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M. A., Redmond J. W., Batley M., Rolfe B. G. Clovers secrete specific phenolic compounds which either stimulate or repress nod gene expression in Rhizobium trifolii. EMBO J. 1987 May;6(5):1173–1179. doi: 10.1002/j.1460-2075.1987.tb02351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S., Abe M. Scanning Electron Microscopy of Rhizobium trifolii Infection Sites on Root Hairs of White Clover. Appl Environ Microbiol. 1980 Dec;40(6):1094–1099. doi: 10.1128/aem.40.6.1094-1099.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak E. M., Urbano M. R., Dazzo F. B. Growth-phase-dependent immunodeterminants of Rhizobium trifolii lipopolysaccharide which bind trifoliin A, a white clover lectin. J Bacteriol. 1981 Nov;148(2):697–711. doi: 10.1128/jb.148.2.697-711.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell D. H., Morales V. M., Umali-Garcia M. Pectolytic enzymes in Rhizobium. Appl Environ Microbiol. 1978 Jan;35(1):210–213. doi: 10.1128/aem.35.1.210-213.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Sato T., Yura T. Synthesis and assembly of the membrane proteins in E. coli. Cell. 1977 Jul;11(3):551–559. doi: 10.1016/0092-8674(77)90073-3. [DOI] [PubMed] [Google Scholar]
- LJUNGGREN H., FAHRAEUS G. The role of polygalacturonase in root-hair invasion by nodule bacteria. J Gen Microbiol. 1961 Nov;26:521–528. doi: 10.1099/00221287-26-3-521. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Martinez-Molina E., Morales V. M., Hubbell D. H. Hydrolytic enzyme production by Rhizobium. Appl Environ Microbiol. 1979 Dec;38(6):1186–1188. doi: 10.1128/aem.38.6.1186-1188.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C. A., Hubbell D. H. Ultrastructure of Rhizobium-induced infection threads in clover root hairs. Appl Microbiol. 1975 Dec;30(6):1003–1009. doi: 10.1128/am.30.6.1003-1009.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares A., Montoya E., Olivares J. Induction of polygalacturonase production in legume roots as a consequence of extrachromosomal DNA carried by Rhizobium meliloti. Microbios. 1978;21(83):33–39. [PubMed] [Google Scholar]
- Ridge R. W., Rolfe B. G. Rhizobium sp. Degradation of Legume Root Hair Cell Wall at the Site of Infection Thread Origin. Appl Environ Microbiol. 1985 Sep;50(3):717–720. doi: 10.1128/aem.50.3.717-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried J. L., Collmer A. Activity stain for rapid characterization of pectic enzymes in isoelectric focusing and sodium dodecyl sulfate-polyacrylamide gels. Appl Environ Microbiol. 1985 Sep;50(3):615–622. doi: 10.1128/aem.50.3.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Palenzuela P., Burr T. J., Collmer A. Polygalacturonase is a virulence factor in Agrobacterium tumefaciens biovar 3. J Bacteriol. 1991 Oct;173(20):6547–6552. doi: 10.1128/jb.173.20.6547-6552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAHLMAN K. AN ELECTRON MICROSCOPE STUDY OF ROOT-HAIR INFECTION BY RHIZOBIUM. J Gen Microbiol. 1963 Dec;33:425–427. doi: 10.1099/00221287-33-3-425. [DOI] [PubMed] [Google Scholar]
- Waffenschmidt S., Jaenicke L. Assay of reducing sugars in the nanomole range with 2,2'-bicinchoninate. Anal Biochem. 1987 Sep;165(2):337–340. doi: 10.1016/0003-2697(87)90278-8. [DOI] [PubMed] [Google Scholar]