Abstract
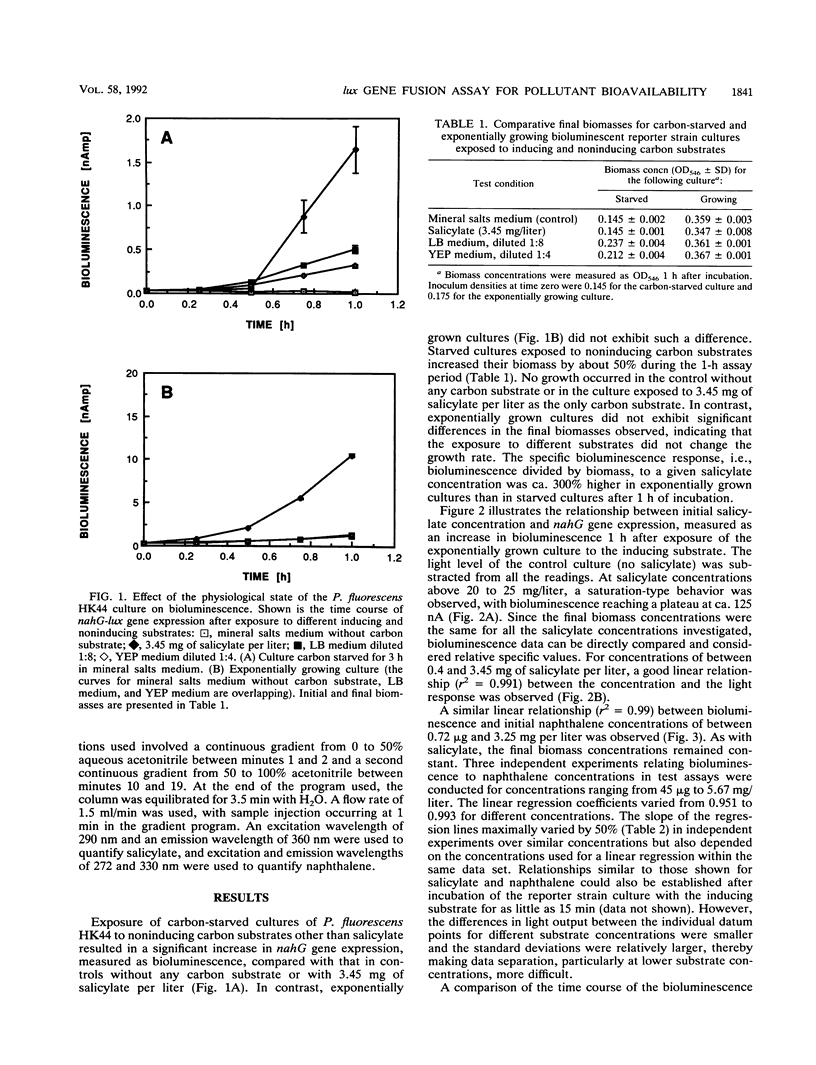
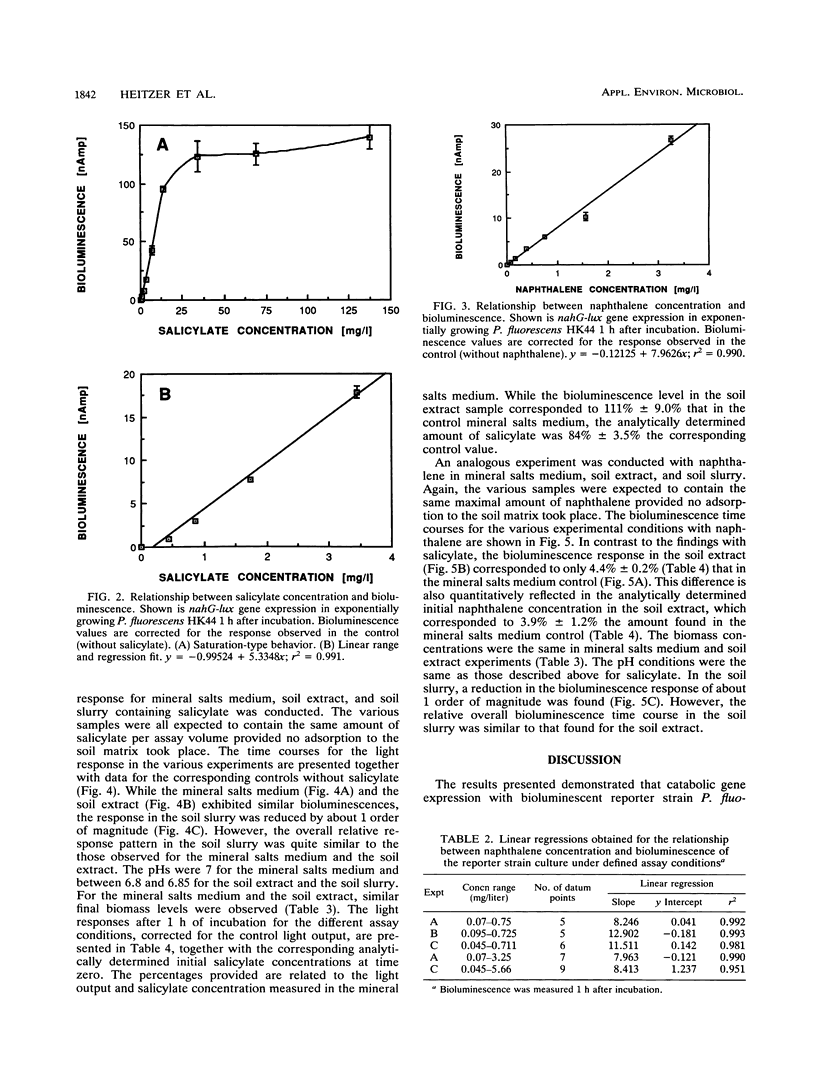
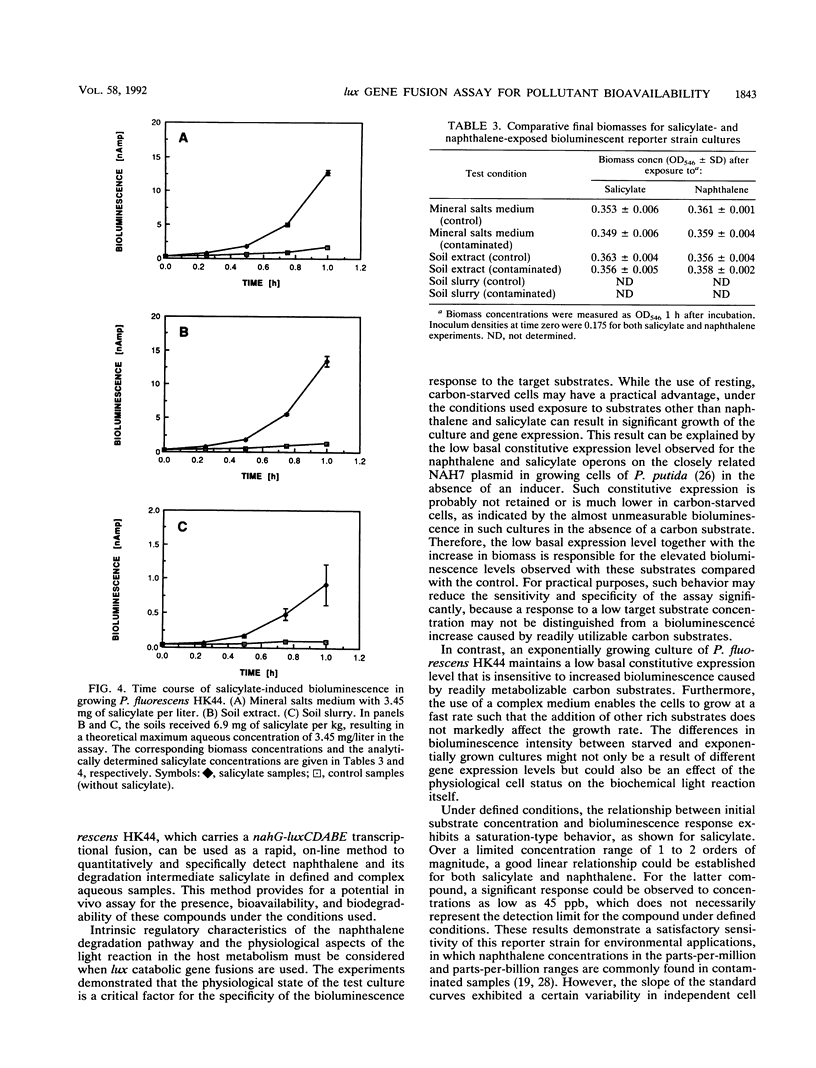
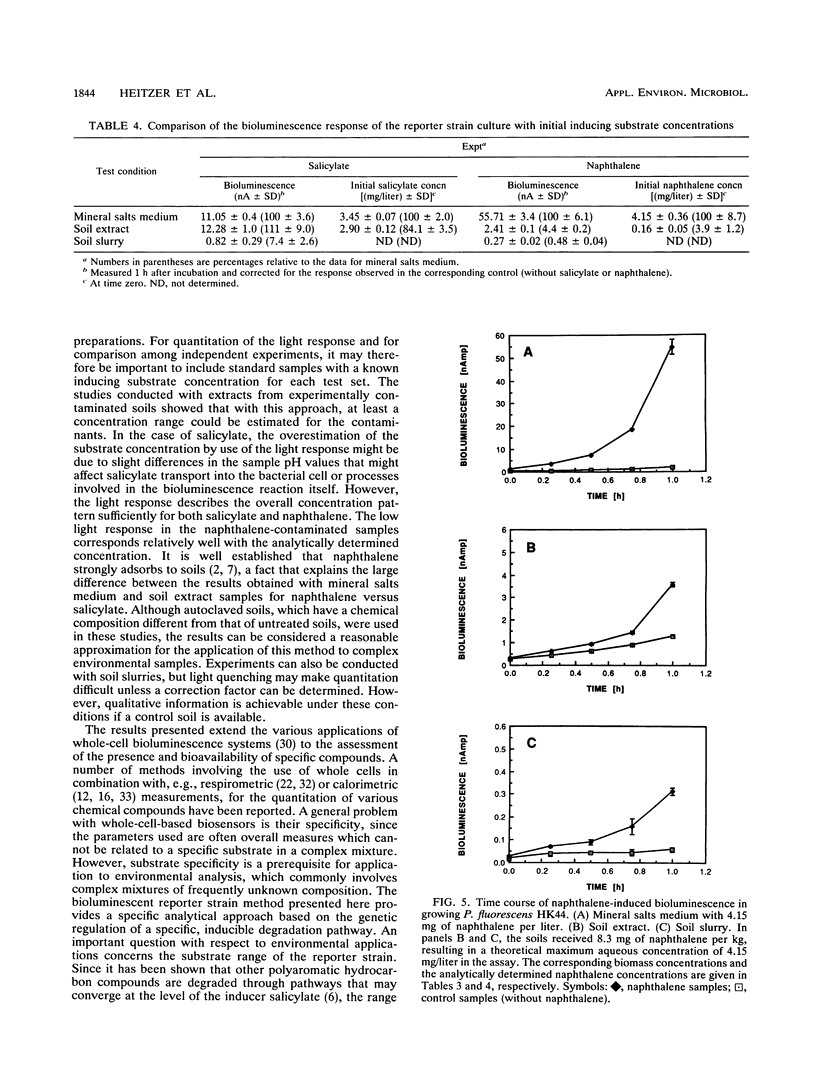
A bioassay was developed and standardized for the rapid, specific, and quantitative assessment of naphthalene and salicylate bioavailability by use of bioluminescence monitoring of catabolic gene expression. The bioluminescent reporter strain Pseudomonas fluorescens HK44, which carries a transcriptional nahG-luxCDABE fusion for naphthalene and salicylate catabolism, was used. The physiological state of the reporter cultures as well as the intrinsic regulatory properties of the naphthalene degradation operon must be taken into account to obtain a high specificity at low target substrate concentrations. Experiments have shown that the use of exponentially growing reporter cultures has advantages over the use of carbon-starved, resting cultures. In aqueous solutions for both substrates, naphthalene and salicylate, linear relationships between initial substrate concentration and bioluminescence response were found over concentration ranges of 1 to 2 orders of magnitude. Naphthalene could be detected at a concentration of 45 ppb. Studies conducted under defined conditions with extracts and slurries of experimentally contaminated sterile soils and identical uncontaminated soil controls demonstrated that this method can be used for specific and quantitative estimations of target pollutant presence and bioavailability in soil extracts and for specific and qualitative estimations of napthalene in soil slurries.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burlage R. S., Sayler G. S., Larimer F. Monitoring of naphthalene catabolism by bioluminescence with nah-lux transcriptional fusions. J Bacteriol. 1990 Sep;172(9):4749–4757. doi: 10.1128/jb.172.9.4749-4757.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi O. A., Stewart G. S., Ulitzur S., Kuhn J. Use of bacterial luciferase to establish a promoter probe vehicle capable of nondestructive real-time analysis of gene expression in Bacillus spp. J Bacteriol. 1987 May;169(5):2165–2170. doi: 10.1128/jb.169.5.2165-2170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Simon M., Silverman M. Measuring gene expression with light. Science. 1985 Mar 15;227(4692):1345–1347. doi: 10.1126/science.2983423. [DOI] [PubMed] [Google Scholar]
- Erickson J. W., Vaughn V., Walter W. A., Neidhardt F. C., Gross C. A. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1987 Jul;1(5):419–432. doi: 10.1101/gad.1.5.419. [DOI] [PubMed] [Google Scholar]
- Hastings J. W., Potrikus C. J., Gupta S. C., Kurfürst M., Makemson J. C. Biochemistry and physiology of bioluminescent bacteria. Adv Microb Physiol. 1985;26:235–291. doi: 10.1016/s0065-2911(08)60398-7. [DOI] [PubMed] [Google Scholar]
- Heitzer A., Mason C. A., Snozzi M., Hamer G. Some effects of growth conditions on steady state and heat shock induced htpG gene expression in continuous cultures of Escherichia coli. Arch Microbiol. 1990;155(1):7–12. doi: 10.1007/BF00291266. [DOI] [PubMed] [Google Scholar]
- Jain R. K., Burlage R. S., Sayler G. S. Methods for detecting recombinant DNA in the environment. Crit Rev Biotechnol. 1988;8(1):33–84. doi: 10.3109/07388558809150537. [DOI] [PubMed] [Google Scholar]
- King J. M., Digrazia P. M., Applegate B., Burlage R., Sanseverino J., Dunbar P., Larimer F., Sayler G. S. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1990 Aug 17;249(4970):778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- Mattiasson B., Larsson P. O., Mosbach K. The microbe thermistor. Nature. 1977 Aug 11;268(5620):519–520. doi: 10.1038/268519a0. [DOI] [PubMed] [Google Scholar]
- Meighen E. A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991 Mar;55(1):123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middaugh D. P., Mueller J. G., Thomas R. L., Lantz S. E., Hemmer M. H., Brooks G. T., Chapman P. J. Detoxification of pentachlorophenol and creosote contaminated groundwater by physical extraction: chemical and biological assessment. Arch Environ Contam Toxicol. 1991 Aug;21(2):233–244. doi: 10.1007/BF01055342. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Schauer A., Ranes M., Santamaria R., Guijarro J., Lawlor E., Mendez C., Chater K., Losick R. Visualizing gene expression in time and space in the filamentous bacterium Streptomyces coelicolor. Science. 1988 May 6;240(4853):768–772. doi: 10.1126/science.3363358. [DOI] [PubMed] [Google Scholar]
- Schell M. A. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene. 1985;36(3):301–309. doi: 10.1016/0378-1119(85)90185-4. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Beckwith J. R. Uses of lac fusions for the study of biological problems. Microbiol Rev. 1985 Dec;49(4):398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. S. In vivo bioluminescence: new potentials for microbiology. Lett Appl Microbiol. 1990 Jan;10(1):1–8. doi: 10.1111/j.1472-765x.1990.tb00082.x. [DOI] [PubMed] [Google Scholar]
- Stucki G., Alexander M. Role of dissolution rate and solubility in biodegradation of aromatic compounds. Appl Environ Microbiol. 1987 Feb;53(2):292–297. doi: 10.1128/aem.53.2.292-297.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodzinski R. S., Bertolini D. Physical state in which naphthalene and bibenzyl are utilized by bacteria. Appl Microbiol. 1972 Jun;23(6):1077–1081. doi: 10.1128/am.23.6.1077-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodzinski R. S., Coyle J. E. Physical state of phenanthrene for utilization by bacteria. Appl Microbiol. 1974 Jun;27(6):1081–1084. doi: 10.1128/am.27.6.1081-1084.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk C. P., Cai Y., Panoff J. M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Serdar C. M. Genetics of naphthalene catabolism in pseudomonads. Crit Rev Microbiol. 1988;15(3):247–268. doi: 10.3109/10408418809104459. [DOI] [PubMed] [Google Scholar]
- al-Bashir B., Cseh T., Leduc R., Samson R. Effect of soil/contaminant interactions on the biodegradation of naphthalene in flooded soil under denitrifying conditions. Appl Microbiol Biotechnol. 1990 Dec;34(3):414–419. doi: 10.1007/BF00170071. [DOI] [PubMed] [Google Scholar]



