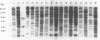Abstract
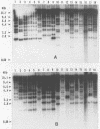
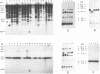
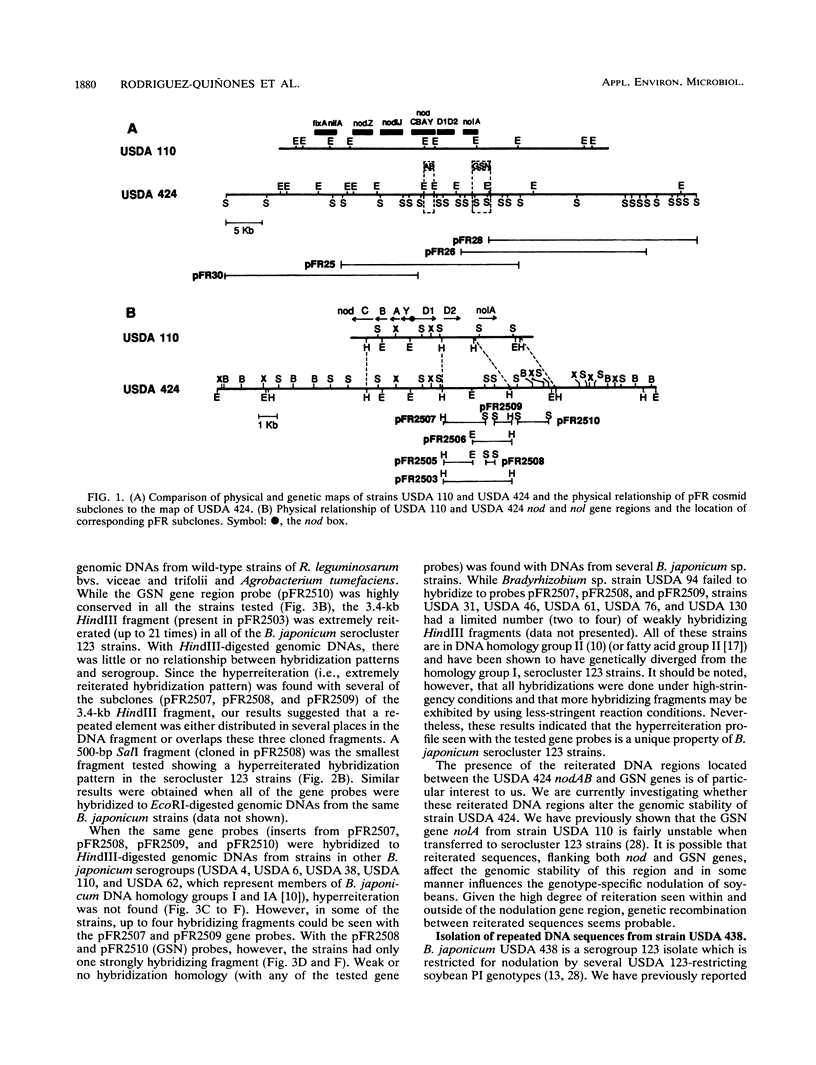
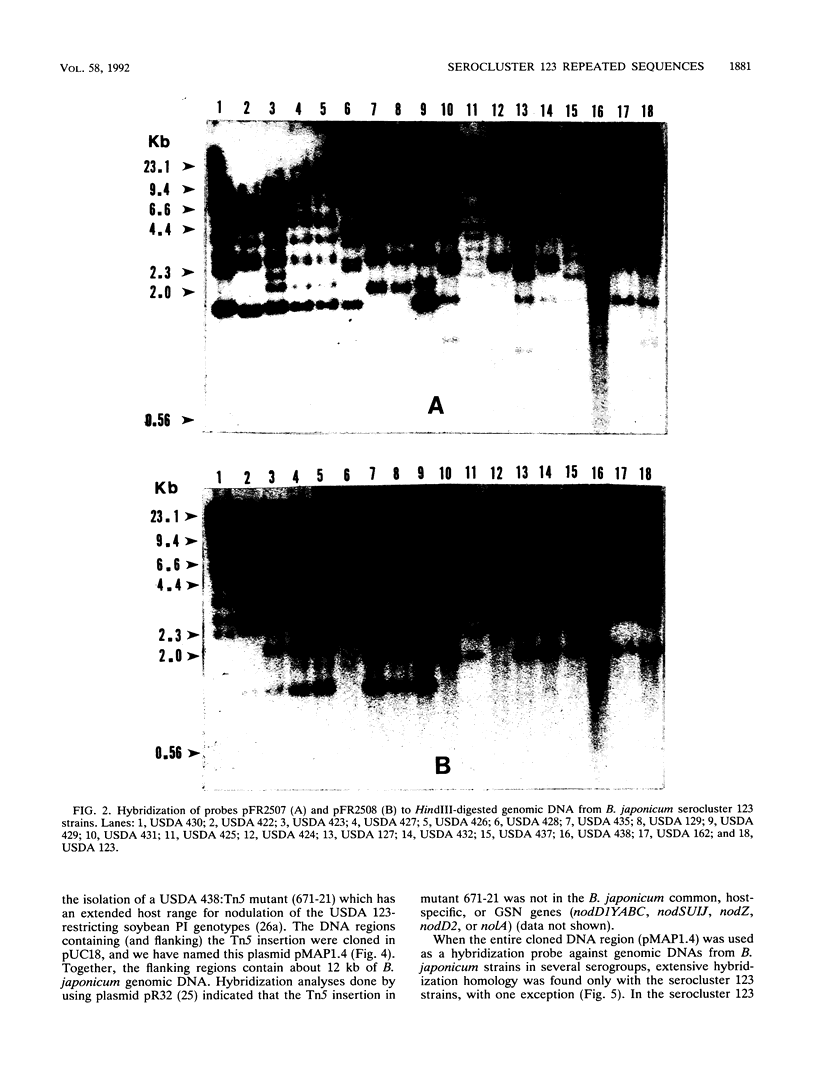
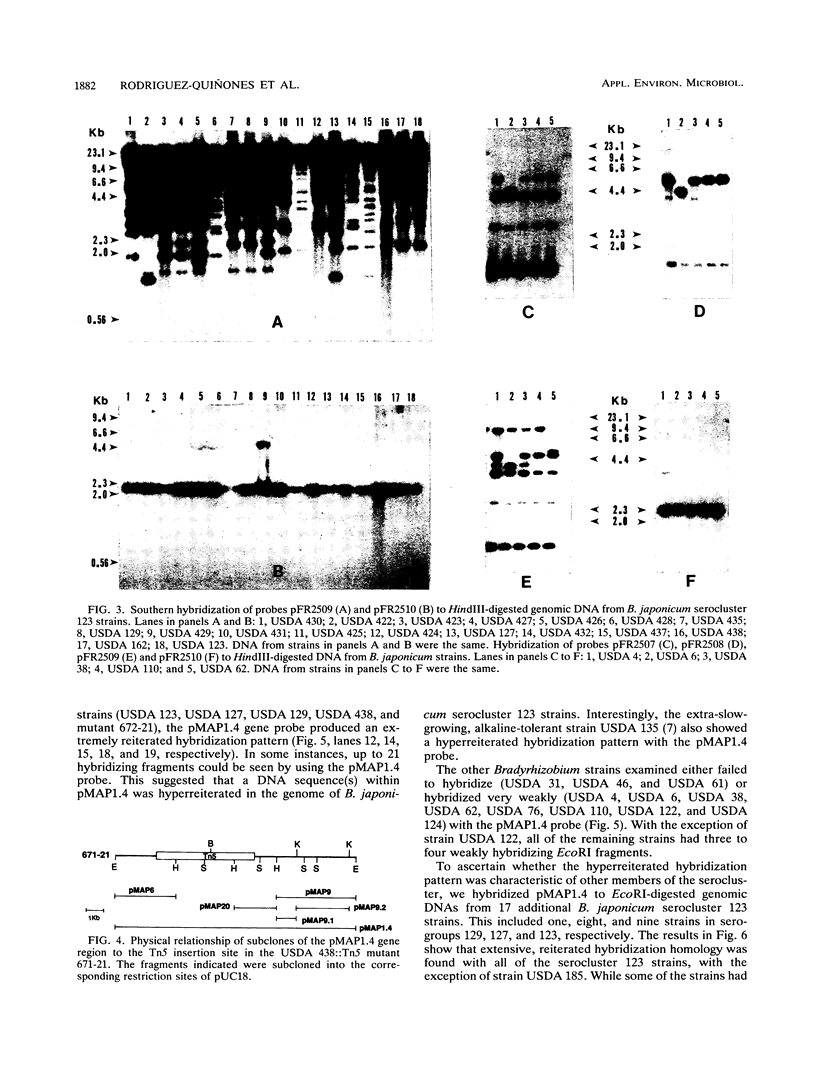
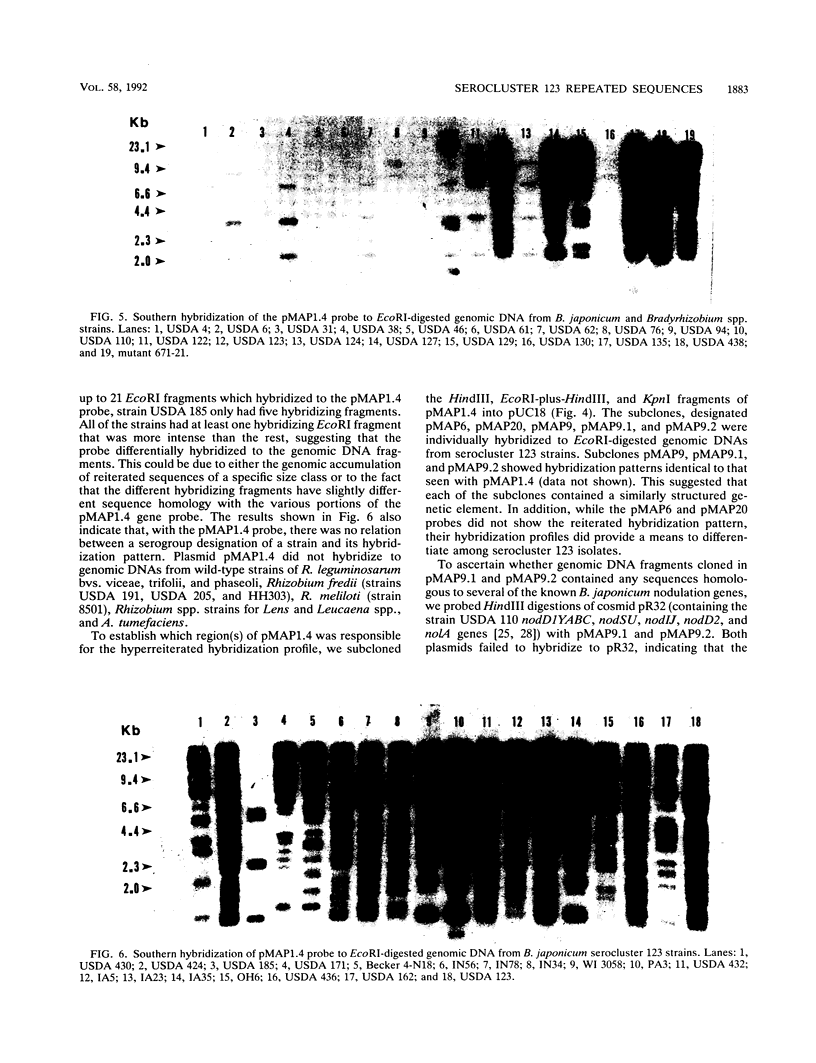
We have identified and cloned two DNA regions which are highly reiterated in Bradyrhizobium japonicum serocluster 123 strains. While one of the reiterated DNA regions, pFR2503, is closely linked to the B. japonicum common and genotype-specific nodulation genes in strain USDA 424, the other, pMAP9, is located next to a Tn5 insertion site in a host-range extension mutant of B. japonicum USDA 438. The DNA cloned in pFR2503 and pMAP9 are reiterated 18 to 21 times, respectively, in the genomes of B. japonicum serocluster 123 strains. Gene probes from the reiterated regions share sequence homology, failed to hybridize (or hybridized poorly) to genomic DNA from other B. japonicum and Bradyrhizobium spp. strains, and did not hybridize to DNA from Rhizobium meliloti, Rhizobium fredii, Rhizobium leguminosarum biovars trifolii, phaseoli, and viceae, or Agrobacterium tumefacians. The restriction fragment length polymorphism hybridization profiles obtained by using these gene probes are useful for discriminating among serologically related B. japonicum serocluster 123 strains.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banfalvi Z., Nieuwkoop A., Schell M., Besl L., Stacey G. Regulation of nod gene expression in Bradyrhizobium japonicum. Mol Gen Genet. 1988 Nov;214(3):420–424. doi: 10.1007/BF00330475. [DOI] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- DATE R. A., DECKER A. M. MINIMAL ANTIGENIC CONSTITUTION OF 28 STRAINS OF RHIZOBIUM JAPONICUM. Can J Microbiol. 1965 Feb;11:1–8. doi: 10.1139/m65-001. [DOI] [PubMed] [Google Scholar]
- Dusha I., Kovalenko S., Banfalvi Z., Kondorosi A. Rhizobium meliloti insertion element ISRm2 and its use for identification of the fixX gene. J Bacteriol. 1987 Apr;169(4):1403–1409. doi: 10.1128/jb.169.4.1403-1409.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttfert M., Lamb J. W., Gasser R., Semenza J., Hennecke H. Mutational analysis of the Bradyrhizobium japonicum common nod genes and further nod box-linked genomic DNA regions. Mol Gen Genet. 1989 Feb;215(3):407–415. doi: 10.1007/BF00427037. [DOI] [PubMed] [Google Scholar]
- Hahn M., Hennecke H. Mapping of a Bradyrhizobium japonicum DNA Region Carrying Genes for Symbiosis and an Asymmetric Accumulation of Reiterated Sequences. Appl Environ Microbiol. 1987 Sep;53(9):2247–2252. doi: 10.1128/aem.53.9.2247-2252.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluza K., Hahn M., Hennecke H. Repeated sequences similar to insertion elements clustered around the nif region of the Rhizobium japonicum genome. J Bacteriol. 1985 May;162(2):535–542. doi: 10.1128/jb.162.2.535-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney B., Staskawicz B. J. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J Bacteriol. 1990 Jan;172(1):143–148. doi: 10.1128/jb.172.1.143-148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser H. H., Cregan P. B. Nodulation and Competition for Nodulation of Selected Soybean Genotypes among Bradyrhizobium japonicum Serogroup 123 Isolates. Appl Environ Microbiol. 1987 Nov;53(11):2631–2635. doi: 10.1128/aem.53.11.2631-2635.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H., Kuykendall L. D., Shah K. S., Keister D. L. Induction of Symbiotically Defective Auxotrophic Mutants of Rhizobium fredii HH303 by Transposon Mutagenesis. Appl Environ Microbiol. 1988 Feb;54(2):423–427. doi: 10.1128/aem.54.2.423-427.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Lewis-Henderson W. R., Djordjevic M. A. A cultivar-specific interaction between Rhizobium leguminosarum bv. trifolii and subterranean clover is controlled by nodM, other bacterial cultivar specificity genes, and a single recessive host gene. J Bacteriol. 1991 May;173(9):2791–2799. doi: 10.1128/jb.173.9.2791-2799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLafferty M. A., Harcus D. R., Hewlett E. L. Nucleotide sequence and characterization of a repetitive DNA element from the genome of Bordetella pertussis with characteristics of an insertion sequence. J Gen Microbiol. 1988 Aug;134(8):2297–2306. doi: 10.1099/00221287-134-8-2297. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Priefer U. B., Burkardt H. J., Klipp W., Pühler A. ISR1: an insertion element isolated from the soil bacterium Rhizobium lupini. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):87–91. doi: 10.1101/sqb.1981.045.01.016. [DOI] [PubMed] [Google Scholar]
- Riley M., Anilionis A. Evolution of the bacterial genome. Annu Rev Microbiol. 1978;32:519–560. doi: 10.1146/annurev.mi.32.100178.002511. [DOI] [PubMed] [Google Scholar]
- Russell P., Schell M. G., Nelson K. K., Halverson L. J., Sirotkin K. M., Stacey G. Isolation and characterization of the DNA region encoding nodulation functions in Bradyrhizobium japonicum. J Bacteriol. 1985 Dec;164(3):1301–1308. doi: 10.1128/jb.164.3.1301-1308.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Long S. R., Meade H. M., van den Bos R. C., Ausubel F. M. ISRm1: A Rhizobium meliloti insertion sequence that transposes preferentially into nitrogen fixation genes. J Mol Appl Genet. 1982;1(5):405–418. [PubMed] [Google Scholar]
- Sadowsky M. J., Bohlool B. B., Keyser H. H. Serological Relatedness of Rhizobium fredii to Other Rhizobia and to the Bradyrhizobia. Appl Environ Microbiol. 1987 Aug;53(8):1785–1789. doi: 10.1128/aem.53.8.1785-1789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Cregan P. B., Gottfert M., Sharma A., Gerhold D., Rodriguez-Quinones F., Keyser H. H., Hennecke H., Stacey G. The Bradyrhizobium japonicum nolA gene and its involvement in the genotype-specific nodulation of soybeans. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):637–641. doi: 10.1073/pnas.88.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Tully R. E., Cregan P. B., Keyser H. H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl Environ Microbiol. 1987 Nov;53(11):2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Zidwick M. J., Abebe H. M. Bradyrhizobium japonicum Serocluster 123 and Diversity among Member Isolates. Appl Environ Microbiol. 1986 Jun;51(6):1212–1215. doi: 10.1128/aem.51.6.1212-1215.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. J., Ames G. F., Smith N. H., Robinson E. C., Higgins C. F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984 Jul;37(3):1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Wheatcroft R., Watson R. J. A Positive Strain Identification Method for Rhizobium meliloti. Appl Environ Microbiol. 1988 Feb;54(2):574–576. doi: 10.1128/aem.54.2.574-576.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]