Abstract
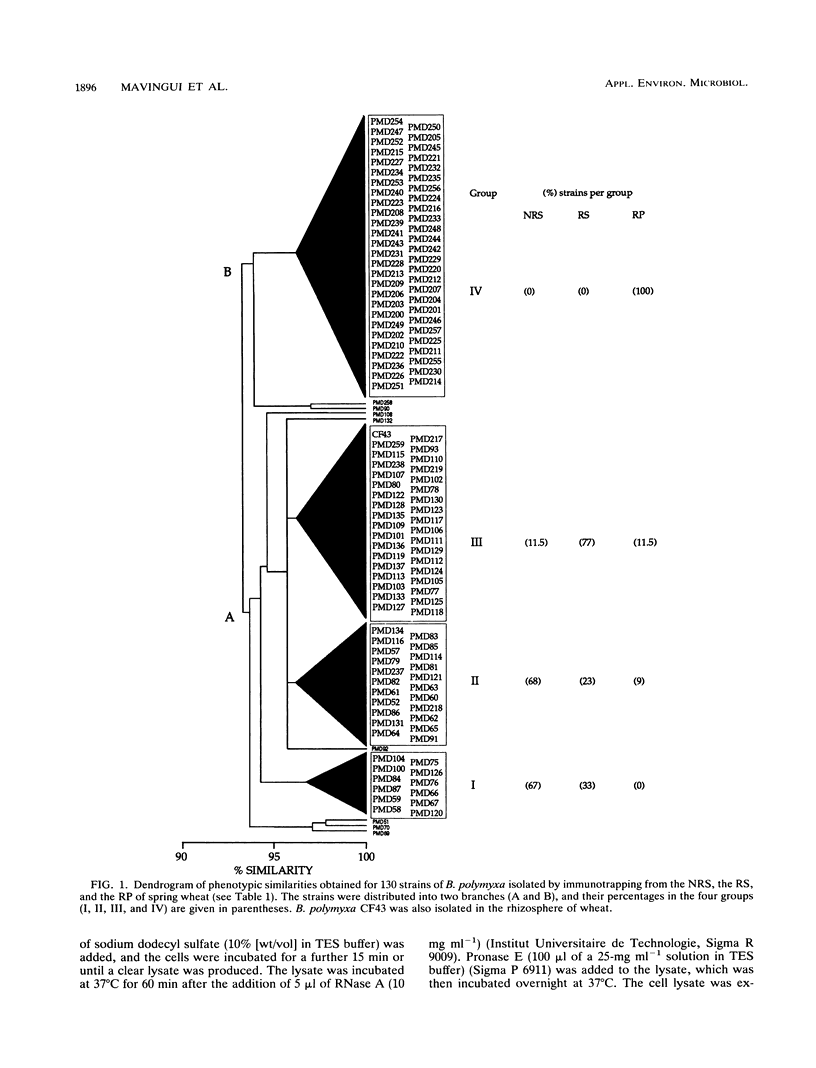
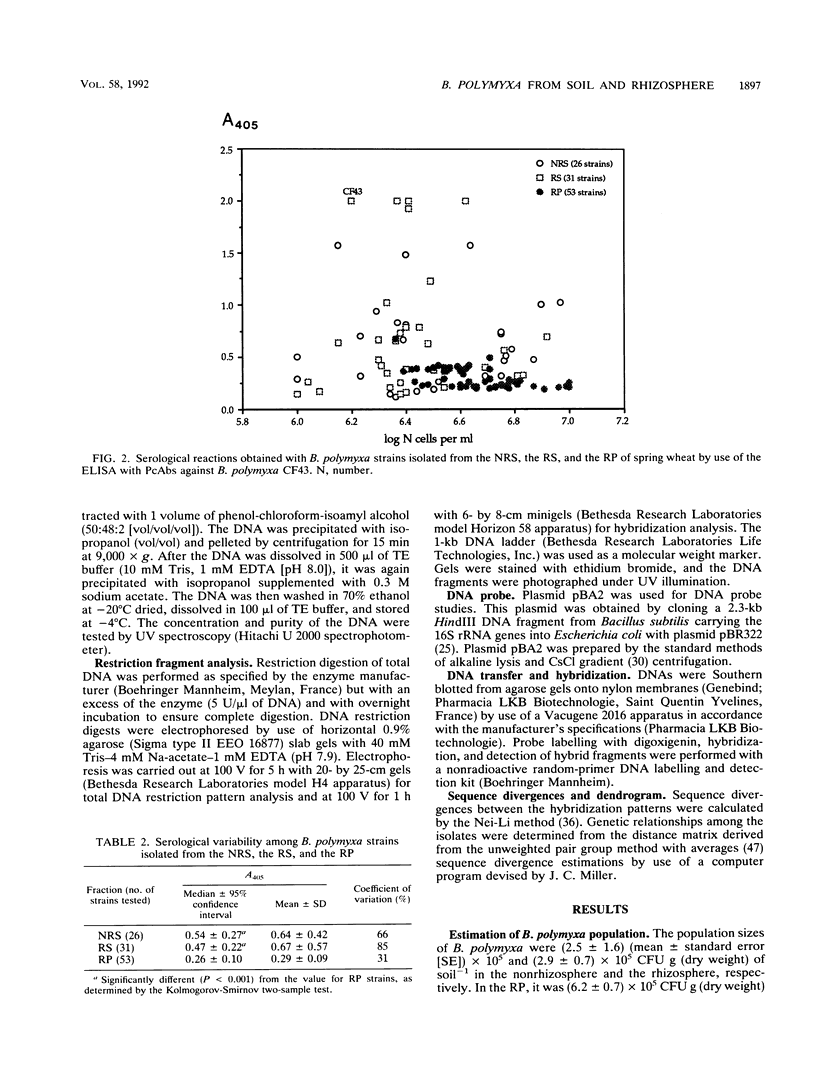
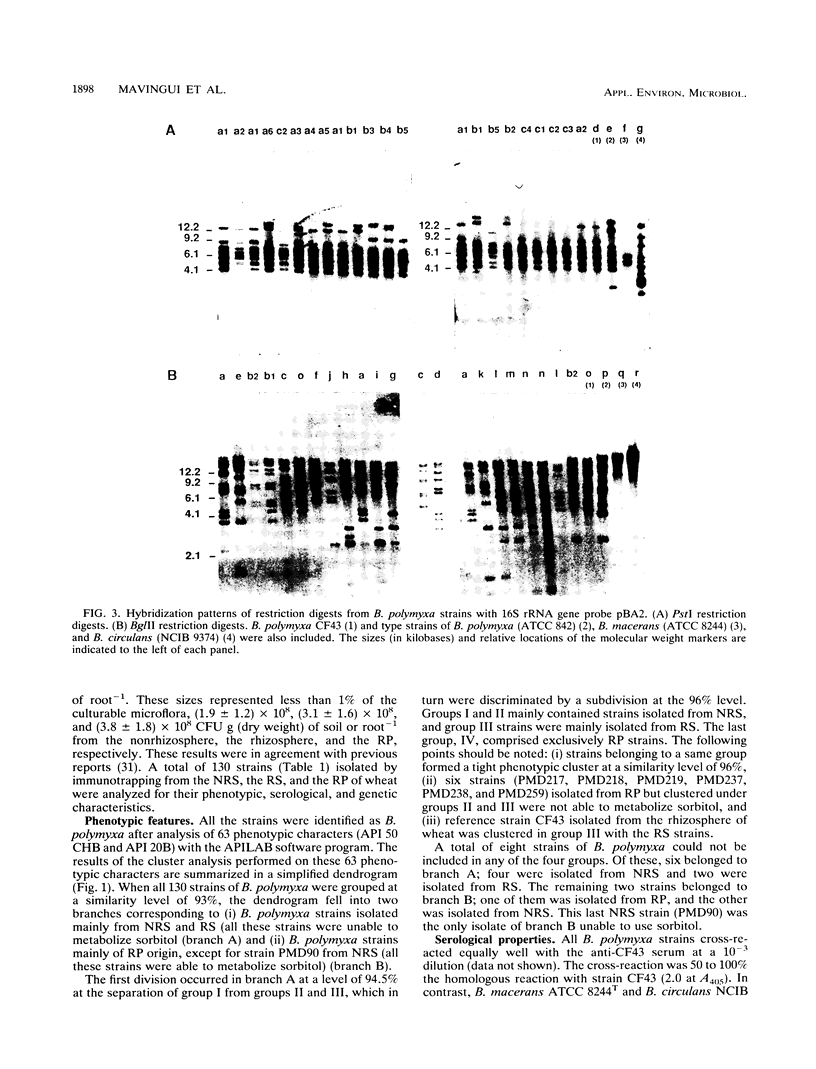
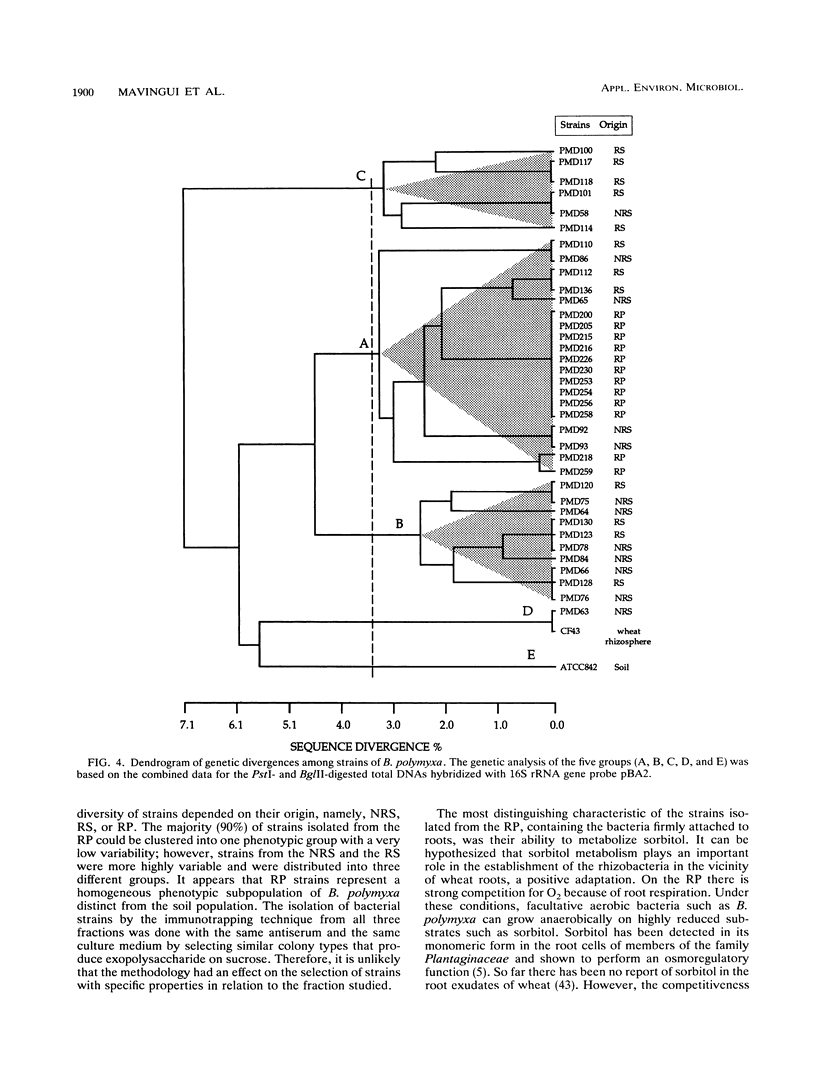
Diversity among 130 strains of Bacillus polymyxa was studied; the bacteria were isolated by immunotrapping from nonrhizosphere soil (32 strains), rhizosphere soil (38 strains), and the rhizoplane (60 strains) of wheat plantlets growing in a growth chamber. The strains were characterized phenotypically by 63 auxanographic (API 50 CHB and API 20B strips) and morphological features, serologically by an enzyme-linked immunosorbent assay, and genetically by restriction fragment length polymorphism (RFLP) profiles of total DNA in combination with hybridization patterns obtained with an rRNA gene probe. Cluster analysis of phenotypic characters by the unweighted pair group method with averages indicated four groups at a similarity level of 93%. Clustering of B. polymyxa strains from the various fractions showed that the strains isolated from nonrhizosphere soil fell into two groups (I and II), while the third group (III) mainly comprised strains isolated from rhizosphere soil. The last group (IV) included strains isolated exclusively from the rhizoplane. Strains belonging to a particular group exhibited a similarity level of 96%. Serological properties revealed a higher variability among strains isolated from nonrhizosphere and rhizosphere soil than among rhizoplane strains. RFLP patterns also revealed a greater genetic diversity among strains isolated from nonrhizosphere and rhizosphere soil and therefore could not be clearly grouped. The RFLP patterns of sorbitol-positive strains isolated from the rhizoplane were identical. These results indicate that diversity within populations of B. polymyxa isolated from nonrhizosphere and rhizosphere soil is higher than that of B. polymyxa isolated from the rhizoplane. It therefore appears that wheat roots may select a specific subpopulation from the soil B. polymyxa population.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankenbauer R. G., Nester E. W. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990 Nov;172(11):6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A. K., McCarty S. C., Atlas R. M. Detection of coliform bacteria and Escherichia coli by multiplex polymerase chain reaction: comparison with defined substrate and plating methods for water quality monitoring. Appl Environ Microbiol. 1991 Aug;57(8):2429–2432. doi: 10.1128/aem.57.8.2429-2432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIES S. N. The serology of Bacillus polymyxa. J Gen Microbiol. 1951 Oct;5(4):807–816. doi: 10.1099/00221287-5-4-807. [DOI] [PubMed] [Google Scholar]
- Graham J. P., Istock C. A. Gene exchange and natural selection cause Bacillus subtilis to evolve in soil culture. Science. 1979 May 11;204(4393):637–639. doi: 10.1126/science.107592. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- HINO S., WILSON P. W. Nitrogen fixation by a facultative bacillus. J Bacteriol. 1958 Apr;75(4):403–408. doi: 10.1128/jb.75.4.403-408.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hranitzky K. W., Larson A. D., Ragsdale D. W., Siebeling R. J. Isolation of O1 serovars of Vibrio cholerae from water by serologically specific method. Science. 1980 Nov 28;210(4473):1025–1026. doi: 10.1126/science.7434013. [DOI] [PubMed] [Google Scholar]
- Iglesias A., Ceglowski P., Trautner T. A. Plasmid transformation in Bacillus subtilis. Effects of the insertion of Bacillus subtilis rRNA genes into plasmids. Mol Gen Genet. 1983;192(1-2):149–154. doi: 10.1007/BF00327660. [DOI] [PubMed] [Google Scholar]
- Lindberg T., Granhall U. Isolation and characterization of dinitrogen-fixing bacteria from the rhizosphere of temperate cereals and forage grasses. Appl Environ Microbiol. 1984 Oct;48(4):683–689. doi: 10.1128/aem.48.4.683-689.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan N. A., Berkeley R. C. Identification of Bacillus strains using the API system. J Gen Microbiol. 1984 Jul;130(7):1871–1882. doi: 10.1099/00221287-130-7-1871. [DOI] [PubMed] [Google Scholar]
- Lynch J. M. Beneficial interactions between micro-organisms and roots. Biotechnol Adv. 1990;8(2):335–346. doi: 10.1016/0734-9750(90)91069-s. [DOI] [PubMed] [Google Scholar]
- Metzger W. C., Klein D. A., Redente E. F. Bacterial physiological diversity in the rhizosphere of range plants in response to retorted shale stress. Appl Environ Microbiol. 1986 Oct;52(4):765–770. doi: 10.1128/aem.52.4.765-770.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moawad H. A., Ellis W. R., Schmidt E. L. Rhizosphere Response as a Factor in Competition Among Three Serogroups of Indigenous Rhizobium japonicum for Nodulation of Field-Grown Soybeans. Appl Environ Microbiol. 1984 Apr;47(4):607–612. doi: 10.1128/aem.47.4.607-612.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R., McCleskey C. S., Levine M. The Facultative Sporulating Bacteria Producing Gas from Lactose. J Bacteriol. 1937 Feb;33(2):163–183. doi: 10.1128/jb.33.2.163-183.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V., Goksøyr J., Daae F. L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V., Salte K., Sørheim R., Goksøyr J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]