Abstract
The INK4a gene, one of the most frequently disrupted tumor suppressor loci in human cancer, encodes two unrelated proteins, p16INK4a and p19ARF, each of which is capable of inducing cell cycle arrest. Splicing of alternative first exons (1α vs. 1β) to a common second exon within INK4a generates mRNAs in which exon 2 sequences are translated in two different reading frames. One of the products, the cyclin D-dependent kinase inhibitor p16INK4a, is functionally inactivated by mutations or deletions in a wide variety of cancers. However, because many such mutations reside in exon 2, they also affect the alternative reading frame (ARF) protein. To determine whether such mutations disrupt p19ARF function, we introduced naturally occurring missense mutations into mouse INK4a exon 2 sequences and tested mutant p16INK4a and p19ARF proteins for their ability to inhibit cell cycle progression. Six p19ARF point mutants remained fully active in mediating cell cycle arrest in NIH 3T3 fibroblasts, whereas two of the corresponding mutations within p16INK4a resulted in complete loss of activity. Analysis of p19ARF deletion mutants indicated that the unique aminoterminal domain encoded by exon 1β was both necessary and sufficient for inducing G1 arrest. Therefore, cancer-associated mutations within exon 2 of the INK4a gene specifically target p16INK4a, and not p19ARF, for inactivation.
Keywords: INK4a tumor suppressor, cyclin D-dependent kinases
Inactivation of the INK4a locus on human chromosome 9p21 by point mutation or deletion is observed in many cancers, including familial melanomas and a wide variety of sporadic tumors of both children and adults (1–5). Accumulating evidence now suggests that the frequency of involvement of the INK4a locus in human cancers may be second only to that of p53, underscoring its broad importance as a tumor suppressor gene (5, 6). Mice nullizygous for INK4a rapidly develop spontaneous tumors whose time of onset is accelerated by carcinogens (7). Embryo fibroblasts derived from such animals fail to senesce in culture and can be transformed by oncogenic RAS alone, suggesting that the loss of INK4a plays a role in cell immortalization. In agreement, the majority of established mouse cell lines lack a functional INK4a gene (1, 2).
The INK4a locus encodes a cyclin D-dependent kinase inhibitor, p16INK4a, whose overexpression in mammalian cells can induce G1 phase arrest (8, 9). By inhibiting the activities of the cyclin D-dependent kinases, cdk4 and cdk6, p16INK4a prevents phosphorylation of the retinoblastoma protein (pRb), a modification that cancels pRb’s growth-suppressive function and allows entry into S phase. This biochemical pathway involving p16INK4a inhibition of cyclin D-dependent pRb kinase activity is frequently disrupted in tumor cells, either by deletions or inactivating mutations of p16INK4a and pRb (tumor suppressors), or by amplification of cyclin D1 or cdk4 (protooncogenes) (5, 6). An alteration of any one gene in the pathway eliminates selective pressure for changes in the others, so that loss of p16 or pRb, or cyclin D–cdk amplification, are mutually exclusive events in the generation of tumor cells.
Three additional INK4 proteins, p15INK4b, p18INK4c, and p19INK4d, are equally potent inhibitors of cdk4 and cdk6, but so far they have not been implicated in tumor suppression, suggesting that the INK4a locus may be unique in this regard. Although p16INK4a is encoded by three exons (designated 1α, 2, and 3), a surprising feature is that the INK4a locus encodes a second, entirely unrelated protein, p19ARF (10). The N-terminal 64 aa of p19ARF are specified by a unique first exon (exon 1β), while its C-terminal 105 aa are encoded by an alternative reading frame (ARF) in exon 2 (10–13). Overexpression of mouse or human p19ARF in rodent fibroblasts and human tumor cells also induces cell cycle arrest, although, unlike p16INK4a, the biochemical function of the ARF protein is unknown (10, 14). Given that some INK4a inactivating mutations in human tumors, including those associated with familial melanoma, are located within exon 1α (15–17), p16INK4a must contribute to tumor suppression in these cases. However, at least half of the tumor-associated INK4a mutations fall within shared exon 2 sequences, so a contribution of p19ARF to tumorigenesis cannot be precluded. Here, we have evaluated the effects of INK4a exon 2 mutations on the growth-arresting capabilities of both p16INK4a and p19ARF and have attempted to pinpoint the domain(s) of p19ARF that are responsible for its cell cycle inhibitory functions.
MATERIALS AND METHODS
Cell Culture and Ectopic Protein Expression.
Cells were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, and 100 units per ml of penicillin and streptomycin (GIBCO). NIH 3T3 fibroblasts were transfected (18) with 30 μg bicistronic pSRα-murine sarcoma virus-tkCD8 retroviral vector plasmids encoding the T cell CD8 coreceptor under control of the thymidine kinase promoter and containing wild-type or mutant p16INK4a cDNAs regulated by the viral long terminal repeat (19). For expression of p19ARF and its mutant derivatives, human kidney 293T cells (20) were transfected with 15 μg of ecotropic helper virus DNA plus 15 μg of the vector plasmid containing wild-type or mutant ARF cDNA. Cell supernatants containing infectious retroviruses were harvested 36–60 hr posttransfection, pooled on ice, and filtered (0.45 μm). Infections of exponentially growing mouse 3T3-D1 cells (21) in 100-mm diameter culture dishes (4 × 105 cells) were performed at 37°C in a 5% CO2 atmosphere using 2.5 ml of virus-containing supernatants supplemented with 8 μg/ml polybrene (Sigma). After 3–6 hr, 10 ml of fresh medium was added, and the medium was changed the following day. Cells were analyzed by dual-color flow cytometry 48 hr after transfection or infection to determine the DNA content of gated CD8-positive and CD8-negative cells (22).
Construction of Mutants.
Six point mutations were introduced into exon 2-derived INK4a cDNA sequences, which were then recloned into full-length cDNAs specifying either mouse p16INK4a or p19ARF. Mutated sense and antisense oligonucleotides complementary to the target sequence strands were used as primers in two separate PCRs (15–25 cycles from 100 ng DNA template) performed with one of two flanking primers complementary to T3 and T7 sequences in the pBluescript vector (Stratagene). These two PCR products were mixed and reamplified (15 cycles, 200 ng DNA) in a subsequent reaction using T3 and T7 primers. Reactions were performed in 10 mM Tris·HCl (pH 8.4), 50 mM KCl, 1 mM MgCl2, 0.1% gelatin, with 50 μM of each dNTP, 1 μg of each primer, and 0.5 units of Taq DNA polymerase (Stratagene). Each cycle consisted of denaturation at 95°C for 1 min, annealing at 65°C for 45 sec, and extension at 72°C for 2 min. The specific oligonucleotides used to construct each mutant, with the mutated nucleotide highlighted in boldface type, are as follows: (i) 5′-CTACCTTCTCTCGCCCGGTGC (sense) and 5′-GCACCGGGCGAGAGAAGGTAG (antisense) to create Pro-93 → Ser in p19ARF and Ser-70 (with a silent third-base change) in p16INK4a; (ii) 5′-CCGGTGCACAACGCAGCGCG (sense) and 5′-CGCGCTGCGTTGTGCACCGG-3′ (antisense) to create Arg-97 → Gln in p19ARF and Asp-75 → Asn in p16INK4a; (iii) 5′-GAAGGCTTCCTGCTGGTGGTG (sense) and 5′-CACCACCAGCAGGAAGCCTTC (antisense) to create ΔGly-105-His-106 in p19ARF and ΔAsp-83-Thr-84 in p16INK4a; (iv) 5′-AGATTCGAACCGCGAGGACCC (sense) and 5′-GGGTCCTCGCGGTTCGAATCTG (antisense) to create Leu-85 → Pro in p19ARF and Cys-63 → Arg in p16INK4a; (v) 5′-CAGATTCGAACGGCGAGGACCC (sense) and 5′-GGGTCCTCGCCGTTCGAATCTG (antisense) to create Leu-85 → Arg in p19ARF and Cys-63 → Gly in p16INK4a; and (vi) 5′-GGCTGGATGTACGCGATGCCTG (sense) and 5′-CAGGCATCGCGTACATCCAGCC-3′ (antisense) to create Ala-120 → Thr in p19ARF and Val-97 (silent third-base change) in p16INK4a. The final PCR products were digested with BstBI and HincII, subcloned at the expense of wild-type sequences into the full-length cDNAs in a pCRII vector (Invitrogen), and verified by resequencing before insertion into the pSRα retroviral expression vector. Mouse p16 mutant cDNAs were also subcloned into the SmaI site of the pGex-4T-2 vector in-frame with glutathione S-transferase (GST).
Deletion mutants lacking p19ARF C-terminal sequences were generated by plasmid cDNA digestion at unique restriction sites, formation of blunt ends, insertion of an XbaI linker encoding stop codons in three frames (5′-CTAGTCTAGACTAG; New England Biolabs), and recircularization. Sites targeted for insertion of the linker were BspLU11I (N45), EcoNI (N62), and BstBI (N84). The Δ1-62 mutant was constructed by digesting hemagglutinin (HA)-p19ARF cDNA with AgeI and EcoNI, removing the fragment encoding the p19ARF N terminus, blunt-ending the remaining vector fragment, and recircularizating it by self-ligation.
Protein Analysis.
For analysis of p19ARF expression by immunoblotting, infected 3T3-D1 cells were pelleted, frozen on dry ice, and lysed in ice-cold Nonidet P-40 buffer (120 mM NaCl/50 mM Tris·HCl, pH 8.0/0.5% Nonidet P-40). After occasional vortexing and incubation for 1 hr on ice, nuclei and debris were removed by centrifugation at 12,000 rpm in a microfuge for 10 min at 4°C. Protein from 2 × 105 cell equivalents was separated on 12.5% denaturing polyacrylamide gels containing SDS/PAGE and transferred onto nitrocellulose. p19ARF was detected by enhanced chemiluminescence (ECL) according to the manufacturer’s specifications (Amersham) using antibodies that recognize its C terminus (10) or the HA epitope (12CA5 monoclonal antiserum; ICN). For binding assays, cdk4 and cdk6 were transcribed and translated in vitro (TnT kit; Promega) and incubated with GST-p16-coated (wild-type vs. mutants) Sepharose beads, and bound products were analyzed by SDS/PAGE, as described (23).
RESULTS
Approximately half of the tumor-associated mutations within the INK4a locus target exon 2, simultaneously altering the primary structure of both p16INK4a and p19ARF. Presumptive polymorphisms that induce no changes in p16INK4a also lead to predicted amino acid substitutions in p19ARF. To test whether p19ARF, like p16INK4a, contributes to tumorigenesis, we introduced six naturally occurring point mutations into the shared exon 2 coding sequences of the cDNAs encoding mouse p16INK4a and p19ARF. Two of the mutations yielded silent, third-base changes in p16INK4a (Ser-70 and Val-97) but generated Pro-93 → Ser and Ala-120 → Thr substitutions, respectively, within p19ARF (1, 24). Another mutation, Asp-75 → Asn, which occurs frequently in sporadic tumors and was previously shown to inactivate the cdk inhibitory activity of p16INK4a (24–26), was predicted to change Arg-97 of p19ARF to Gln. Conversely, a p16INK4a mutation (Cys-63 → Arg) that has no effect on its activity was introduced into p19ARF, thereby replacing Leu-85 → Pro. Because Cys-63 of p16INK4a is independently mutated to Gly in some cancers (25, 27), we also introduced this mutation into p16INK4a and made the corresponding Leu-85 → Arg mutation in p19ARF. Finally, two aa were deleted from p16 (Asp-83 and Thr-84, designated ΔAsp-83-Thr-84) and p19ARF (ΔGly-105-His-106). These residues lie within a 12-aa string that is 92% identical in mouse and human INK4a and is often mutated in tumors.
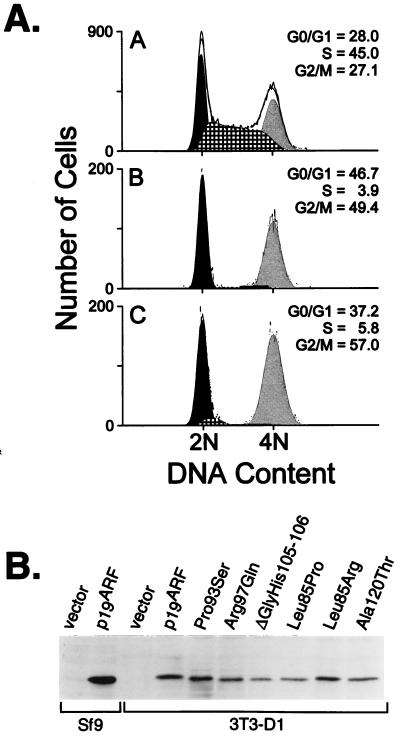
Cells enforced to overexpress the wild-type p19ARF protein arrest in both the G1 and G2 phases of the cell cycle (10). Mutated p19ARF cDNAs were inserted into a bicistronic retroviral expression vector that also encodes the T cell coreceptor, CD8. After transfection of the vector plasmids together with a helper virus into human 293T cells, transiently produced, high-titer viruses released into the culture supernatants were used to infect NIH 3T3 cells engineered to overexpress cyclin D1. This 3T3-D1 derivative was chosen because it undergoes a more significant G2 arrest than parental NIH 3T3 cells in response to enforced p19ARF expression (10). Infected cells were identified by CD8 immunofluorescence, and their DNA content was simultaneously determined after propidium iodide staining using two-color flow cytometry. As opposed to asynchronously replicating cells or controls infected with the CD8 vector alone (Fig. 1A Top), the DNA content of CD8-positive cells expressing wild-type mouse p19ARF showed nearly complete arrest in the G1 and G2 phases of the cell cycle (Fig. 1A Middle). All six p19ARF point and deletion mutants remained fully active in mediating cell cycle arrest; Fig. 1A Bottom shows a typical result with the Pro-93 → Ser mutant. Immunoblotting revealed nearly equivalent expression of the p19ARF mutants in the infected cells (Fig. 1B). Note that p19ARF and p16INK4a are not normally expressed in NIH 3T3 fibroblasts, which have sustained biallelic INK4a deletions.
Figure 1.
Exon-2 mutations have no effect on the growth inhibitory activity of p19ARF. (A) The DNA content of successfully infected NIH 3T3-D1 cells (CD8+) was analyzed 48 hr after infection with a control vector (Top), wild-type p19ARF (Middle), or derivative p19ARF retroviruses containing point mutations within exon 2 sequences (representative mutant Pro-93 → Ser shown, Bottom). All six p19ARF point mutants displayed nearly identical arrest profiles, yielding average cell cycle distributions of 44.2 ± 5.3% G0/G1, 5.7 ± 2.2% S, and 50.1 ± 7.2% G2. By comparison, cells infected with a control vector retrovirus showed cell cycle profiles of 26.2 ± 3.5% G0/G1, 43.9 ± 2% S, and 29.9 ± 4.8% G2/M, averaged from six independent experiments. (B) Expression levels of the mutant proteins compared with wild-type p19ARF were determined by direct Western blot analysis of lysates (2 × 105 cell equivalents per lane) prepared from the infected cells.
The ability of the corresponding mouse p16INK4a mutants to induce G1 arrest in NIH 3T3 fibroblasts and to bind to cdk4 and cdk6 in vitro was also assessed (data not shown). These results, together with those for the ARF mutants, are summarized in Table 1. p16INK4a proteins containing silent Ser-70 or Val-97 mutations behaved indistinguishably from wild-type p16, causing G1 arrest in NIH 3T3 cells and retaining strong cdk4 and cdk6 binding affinity. Likewise, altering Cys-63 to Arg or to Gly did not affect these activities. In contrast, the p16 Asp-75 → Asn and ΔAsp-83-Thr-84 mutants were completely inactive in both assays. Thus, p16INK4a activity was eliminated by nucleotide changes that had no effect on the ability of p19ARF to induce cell cycle arrest. These results suggest that tumor-associated mutations in exon 2 of the INK4a gene specifically target p16 for inactivation.
Table 1.
Analysis of shared exon-2 point mutations within mouse p19ARF and p16INK4a
| p19ARF
|
p16INK4a
|
||
|---|---|---|---|
| Codon mutation | G1 + G2 arrest* | Codon mutation | G1 arrest† |
| Pro93Ser | + | Silent Ser70 | + |
| Arg97Gln | + | Asp75Asn | — |
| ΔGlyHis105–106 | + | ΔAspThr83–84 | — |
| Leu85Pro | + | Cys63Arg | + |
| Leu85Arg | + | Cys63Gly | + |
| Ala120Thr | + | Silent Val97 | + |
Cell cycle distributions were determined for CD8+ NIH 3T3-D1 cells following infection with retroviruses encoding p19ARF mutants and the cell surface marker, CD8. A typical G1 and G2 arrest profile obtained with the mutants is shown in Fig. 1A, with averaged values for the cell cycle distribution listed in the figure legend.
Following transfection with bicistronic plasmids encoding mouse p16 and CD8, CD8+ cells expressing active mouse p16 mutants showed G1 arrest profiles of 86.7 ± 2.6% G0/G1, 9.0 ± 3.5% S phase, and 4.3 ± 1.1% G2/M. Similarly, cells overexpressing wild-type p16INK4a yielded cell cycle distributions of 86.1 ±1.1% G0/G1, 12.0 ± 1.1% S phase, and 1.9 ± 0.1% G2/M. Profiles obtained with empty vector control (69.4 ±1.9% G0/G1, 20.5 ± 3.8% S, 10.1 ± 1.9% G2/M) or inactive mouse p16 mutants (69.3 ± 3.5% G0/G1, 17.6 ± 3.8% S, 13.2 ± 2.1% G2/M) were highly comparable. Mouse p16INK4a mutants were also expressed as GST-fusion proteins and tested for their ability to bind in vitro transcribed and translated cdk4 or cdk6. All mutants that induced G1 arrest efficiently bound cdk4 and cdk6, whereas inactive mutants were incapable of binding either cdk4 or cdk6.
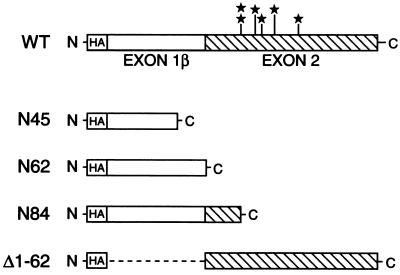
To identify domains within p19ARF that are required for cell cycle arrest, we constructed processive deletion mutants that lacked portions of the N- or C-terminal portions of the protein (Fig. 2). Two mutants were of particular interest because they encoded proteins derived exclusively from either exon 1β (N62) or exon 2 (Δ1-62), respectively. Two other C-terminal deletion mutants were generated, one containing 22 aa of exon 2 in addition to exon 1β (N84) and another encoding only 45 aa of exon 1β, designated N45. Because our antiserum to p19ARF recognizes its distal C terminus, we cloned all wild-type and mutant derivatives downstream of an N-terminal HA epitope tag to facilitate equal detection of the various proteins. We then assayed the ability of each deletion mutant to induce cell cycle arrest in fibroblasts using the same method as that used for the p19ARF point mutants.
Figure 2.
Schematic of p19ARF deletion and point mutants. The N-terminal 64 aa of p19ARF are encoded by exon 1β (open bar), while its C-terminal 105 aa are specified by exon 2 (cross-hatched bar) of the INK4a locus. Point mutations within exon 2 are starred in the wild-type (WT) version of p19ARF, and the deletion mutants are depicted below it. All mutants were tagged with an N-terminal HA epitope to facilitate detection.
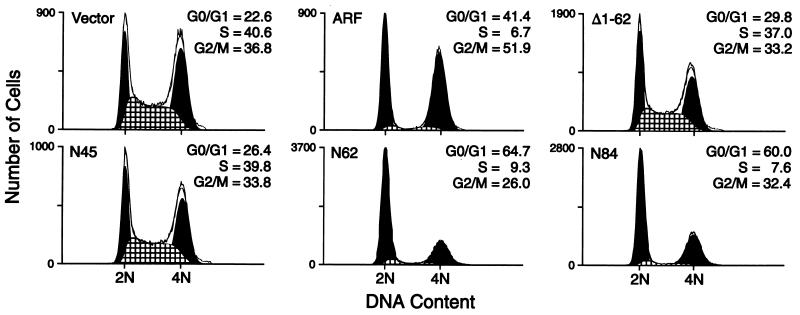
Compared with proliferating cells infected with the control CD8-expressing vector, 3T3-D1 cells infected with wild-type p19ARF retroviruses reproducibly underwent arrest in both G1 and G2 with less than 10% of cells in S phase (Fig. 3). Two mutants which partly or completely lacked exon 1β sequences, N45 and Δ1-62, were inactive and yielded cells with DNA content profiles identical to those of asynchronously proliferating cells. Conversely, mutants retaining exon 1β sequences, N62 and N84, showed almost full activity, inducing arrest predominantly in G1 with a lower percentage in G2 than that seen with the wild-type p19ARF protein.
Figure 3.
Exon-1β sequences specific to p19ARF are necessary and sufficient to induce cell cycle arrest. NIH 3T3-D1 cells were infected with viruses prepared from empty vector, full-length p19ARF (ARF), or deletion mutants, as indicated, and 48 hr after infection the DNA content of CD8+ cells (successfully infected) was measured. The percentage of cells in each phase of the cell cycle is shown for a representative experiment comparing each construct. In three to six independent experiments, the cell cycle fractions obtained for each construct were as follows: vector, 26.2 ± 3.5% G0/G1, 43.9 ± 2% S, and 29.9 ± 4.8% G2/M; WT ARF, 43.9 ± 6.8% G0/G1, 7.6 ± 3.5% S, and 48.5 ± 7.2% G2; N45, 24.4 ± 3% G0/G1, 41.3 ± 1.6% S, and 34.3 ± 1.5% G2/M; N62, 56.9 ± 7.2% G0/G1, 12.9 ± 3% S, and 30.2 ± 4.1% G2; N84, 58.4 ± 6% G0/G1, 10.6 ± 6% S, and 31.0 ± 3.5% G2; and Δ1-62, 27.4 ± 2.7% G0/G1, 38.3 ± 1.6% S, and 34.3 ± 1.9% G2/M.
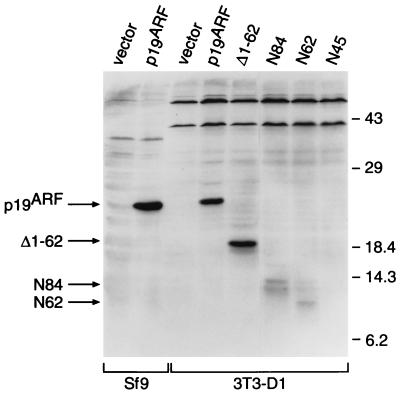
Expression of the mutant proteins was examined by immunoblotting using antiserum against the HA tag (Fig. 4). HA-tagged p19ARF expressed in Sf9 cells was used as a positive control, whereas control vector-infected 3T3-D1 cells again lacked the protein. Although transcription and translation in vitro confirmed that the N45 cDNA encodes an ≈7200-Da polypeptide that was specifically immunoprecipitated by 12CA5 antiserum against the HA epitope (data not shown), we were unable to detect N45 protein in infected 3T3-D1 cells either by immunoblotting (Fig. 4) or by immunofluorescence (data not shown). However, the inactive Δ1-62 mutant was reproducibly expressed at levels equivalent to, or higher than, that of wild-type p19ARF. Even though they retained activity (Fig. 3), the N84 and N62 proteins were detected at much lower levels than that of Δ1-62 (Fig. 4). Together, these results indicate that most p19ARF activity resides in the domain encoded by exon 1β, consistent with the inability of mutations targeted to exon 2 to affect p19ARF activity.
Figure 4.
Expression of p19ARF deletion mutants in NIH 3T3-D1 fibroblasts. Lysates were prepared from 3T3-D1 cells infected with viruses encoding wild-type p19ARF and deletion mutants, shown in Fig. 3, and were analyzed by Western blot analysis. Samples (2 × 105 cell equivalents) were separated by SDS/PAGE and transferred to nitrocellulose, and proteins were detected by ECL using the 12CA5 mAb directed against the HA epitope tag. Lysates from insect Sf9 cells expressing empty vector or HA-tagged p19ARF were used as controls. The sizes of protein molecular weight standards are indicated at the right (kDa), while the positions of the wild-type and mutant p19ARF proteins are indicated with arrows in the left margin.
DISCUSSION
Deletions of the INK4a locus, as well as nearly half of the INK4a mutations detected in human tumors, affect both p16INK4a and p19ARF. The notion that each of these proteins may potentially contribute to tumor suppression stems from the fact that enforced ectopic expression of either can induce cell cycle arrest. We attempted to evaluate whether p19ARF might play a role in tumorigenesis by introducing naturally occurring INK4a exon 2 mutations into cDNAs encoding mouse p19ARF and p16INK4a. Six p19ARF missense mutants remained fully active in blocking cell cycle progression, whereas two of the corresponding mutations in p16INK4a eliminated its ability to bind cdk4 or cdk6 in vitro and to induce G1 arrest. At face value, these results suggest that inactivating mutations within exon 2 of the INK4a locus specifically target p16INK4a and imply in turn that p19ARF is not a tumor suppressor. Consistent with this idea are observations that many tumor-associated mutations truncate the p16INK4a protein but result in less drastic missense mutations in p19ARF (7). In agreement, we found that the p19ARF mutant derived exclusively from exon 2 coding sequences (Δ1-62) was incapable of inhibiting cell cycle progression, despite its high levels of expression, whereas removal of exon 2 coding sequences in the N62 and N84 mutants only marginally reduced their ability to halt the cell cycle. Hence, the majority of p19ARF activity appears to be encoded by its unique exon 1β sequences.
Although the sequence of p16INK4a is selectively targeted in tumors, it still remains unclear as to whether exon 1β of INK4a can contribute to cancer. Tumor-associated deletions in chromosome 9p21 often encompass both exon 1α and 1β, as well as the adjacent gene encoding p15INK4b (28). In some cases of acute lymphocytic leukemia, however, p16INK4a sequences were spared while p15INK4b and exon 1β were specifically deleted (29). Likewise, two human melanoma cell lines, A375 and SK-Mel93, sustained localized homozygous deletions of exon 1β while exon 1α, 2, and 3 of INK4a remained intact allowing expression of p16INK4a transcripts (13). Yet when melanoma-prone kindreds or sporadic melanoma cases were examined, no mutations in exon 1β were identified (13, 30). Rather, most INK4a mutations associated with hereditary melanoma in Dutch and Australian families reside in exon 1α (15–17). A number of tumor-derived lung, bladder, and glioma cell lines were also screened for exon 1β mutations, and none were found (13). It is noteworthy, however, that the latter cell lines did not contain homozygous deletions or mutations involving p16INK4a, indicating that the INK4a locus was not targeted in these cases. Because mice engineered to lack INK4a should have retained exon 1β (7), it is conceivable that an active, truncated ARF protein is produced in these animals. Independent disruption of INK4a exon 1α and 1β in mice should further help to clarify the respective roles of the two proteins in mouse development and cancer.
The hallmark of p19ARF-induced arrest is the redistribution of cells into the two gap phases of the cycle at the expense of cells in S phase and mitosis (10). Although the exact manner by which p19ARF induces arrest remains unclear, the simplest idea is that the overexpressed protein can interfere with a process that is required for both the G1 to S and the G2 to M phase transitions. Viral titers for all of the mutants were reproducibly high, resulting in ≥90% of cells becoming successfully infected. Yet, p19ARF mutants lacking the C terminus were consistently expressed at lower (N62 and N84) or undetectable (N45) levels compared with wild-type p19ARF or the Δ1-62 mutant, so a region encoded by exon 2 may help to stabilize the protein. The lower levels of the N62 and N84 proteins were still sufficient to induce arrest, although cells expressing these mutants were also more prone to accumulate in G1 than G2. If G1 phase cells are more sensitive than G2 cells to the effects of p19ARF, lower levels of expression may favor G1 phase arrest. We think it is unlikely that sequences specifically required for G2 arrest map to the C terminus, because the Δ1-62 mutant was devoid of activity.
We previously detected significantly higher levels of p19ARF in p53-negative cell lines (10). In more recent studies utilizing fibroblasts expressing temperature-sensitive p53 or expressing the p53-inactivating oncoprotein, human papillomavirus E6, we have not observed p53-dependent regulation of p19ARF expression. Instead, p19ARF levels tend to increase as cell strains are progressively passaged in culture (data not shown). By virtue of their establishment, most p53-negative cell lines have been maintained in culture for long periods of time, and this may explain why they express higher levels of p19ARF. When immunoprecipitated from metabolically labeled infected cells, p19ARF specifically associates with several proteins under conditions where the inactive Δ1-62 mutant does not form complexes (data not shown). The identification and characterization of these associated proteins may provide further clues about the role of p19ARF in cell cycle control.
Acknowledgments
We are grateful to Frederique Zindy for performing immunofluorescence to examine expression of p19ARF mutants in fibroblasts, and to Alan Diehl for critical reading of the manuscript. This work was supported in part by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
Footnotes
Abbreviations: ARF, alternative reading frame; pRb, retinoblastoma protein; HA, hemagglutinin; GST, glutathione S-transferase.
References
- 1.Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian S V, Stockert E, Day R S, III, Johnson B E, Skolnick M H. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 2.Nobori T, Miura K, Wu D J, Lois A, Takabayashi K, Carson D A. Nature (London) 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 3.Hirama T, Koeffler H P. Blood. 1995;86:841–854. [PubMed] [Google Scholar]
- 4.Pollock P M, Pearson J V, Hayward N K. Genes Chromosomes Cancer. 1996;15:77–88. doi: 10.1002/(SICI)1098-2264(199602)15:2<77::AID-GCC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 6.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 7.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 8.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 9.Serrano M, Gomez-Lahoz E, DePinho R A, Beach D, Bar-Sagi D. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 10.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 11.Duro D, Bernard O, Della Valle V, Berger R, Larsen C-J. Oncogene. 1995;11:21–29. [PubMed] [Google Scholar]
- 12.Mao L, Merlo A, Bedi G, Shapiro G I, Edwards C D, Rollins B J, Sidransky D. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- 13.Stone S, Jiang P, Dayanauth P, Tavtigian S V, Katcher H, Parry D, Peters G, Kamb A. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- 14.Liggett W H, Jr, Sewell D A, Rocco J, Ahrendt S A, Koch W, Sidransky D. Cancer Res. 1996;56:4119–4123. [PubMed] [Google Scholar]
- 15.Gruis N A, van der Velden P A, Sandkuijl L A, Prins D E, Weaver-Feldhaus J, Kamb A, Bergman W, Frants R. Nat Genet. 1995;10:351–353. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- 16.Holland E A, Beaton S C, Becker T M, Grulet O M C, Peters B A, Rizos H, Kefford R F, Mann G J. Oncogene. 1995;11:2289–2294. [PubMed] [Google Scholar]
- 17.Walker G J, Hussussian G J, Flores J F, Glendening J M, Haluska F G, Dracopoli N C, Hayward N K, Fountain J W. Hum Genet. 1995;4:1845–1852. doi: 10.1093/hmg/4.10.1845. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller A J, Young J C, Pendergast A M, Pondel M, Landau N R, Littman D R, Witte O N. Mol Cell Biol. 1994;14:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pear W S, NoPan M L, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quelle D E, Ashmun R A, Shurtleff S A, Kato J, Bar-Sagi D, Roussel M F, Sherr C J. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 22.Look A T, Melvin S L, Brown L K, Dockter M E, Roberson P K, Murphy S B. J Clin Invest. 1984;73:1617–1628. doi: 10.1172/JCI111368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quelle D E, Ashmun R A, Hannon G J, Rehberger P A, Trono D, Richter H, Walker C, Beach D, Sherr C J, Serrano M. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 24.Zhang S Y, Klein-Szanto A J, Sauter E R, Shafarenko M, Mitsunaga S, Nobori T, Carson D A, Ridge J A, Goodrow T L. Cancer Res. 1994;54:5050–5053. [PubMed] [Google Scholar]
- 25.Mori T, Miura K, Aoki T, Nishihira T, Mori S, Nakamura Y. Cancer Res. 1994;54:3396–3397. [PubMed] [Google Scholar]
- 26.Koh J, Enders G H, Dynlacht B D, Harlow E. Nature (London) 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 27.Li Y-J, Hoang-Xuan K, Delattre J-Y, Poisson M, Thomas G, Hamelin R. Oncogene. 1995;11:597–600. [PubMed] [Google Scholar]
- 28.Stone S, Dayanauth P, Jiang P, Weaver-Feldhaus J M, Tavtigian S V, Cannon-Albright L, Kamb A. Oncogene. 1995;11:987–991. [PubMed] [Google Scholar]
- 29.Heyman M, Rasool O, Brandter L B, Liu Y, Grander D, Einhorn S, Soderhall S. Blood. 1996;87:1657–1658. [PubMed] [Google Scholar]
- 30.Fitzgerald M G, Harkin D P, Silva-Arrieta S, MacDonald D J, Lucchina L C, Unsal H, O’Neill E, Koh J, Finkelstein D M, Isselbacher K J, Sober A J, Haber D A. Proc Natl Acad Sci USA. 1996;93:8541–8545. doi: 10.1073/pnas.93.16.8541. [DOI] [PMC free article] [PubMed] [Google Scholar]