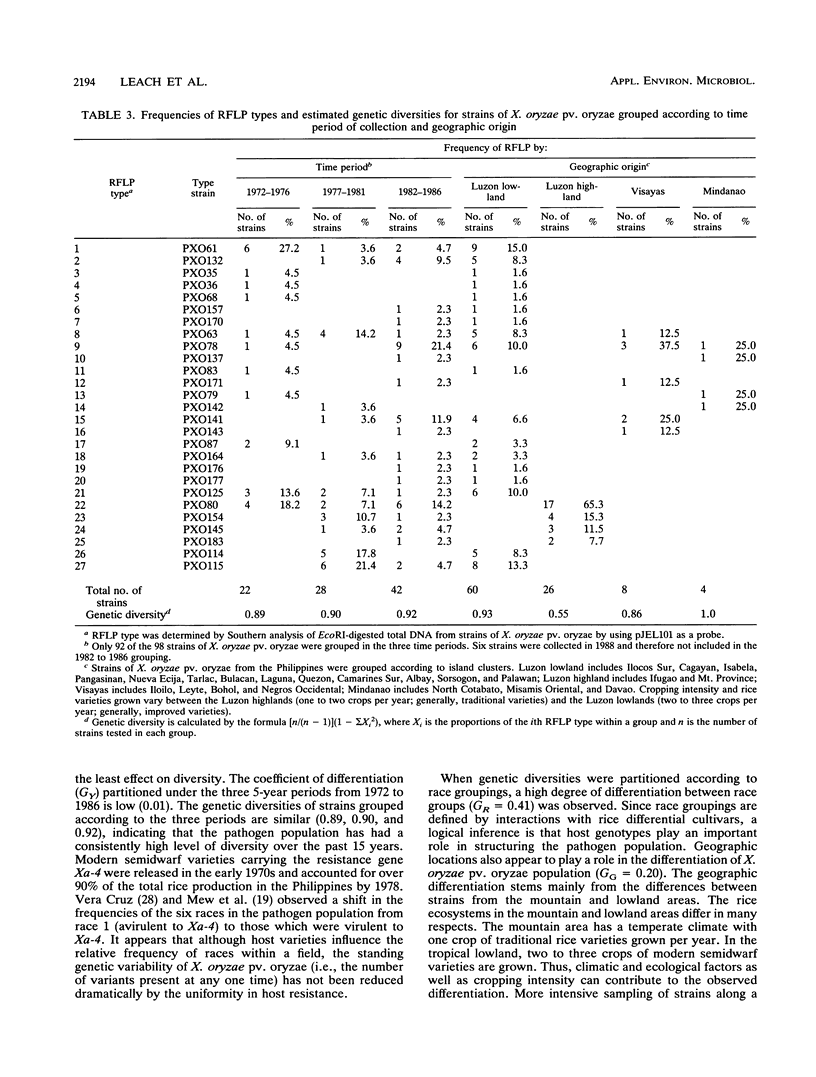
Abstract
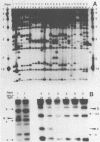
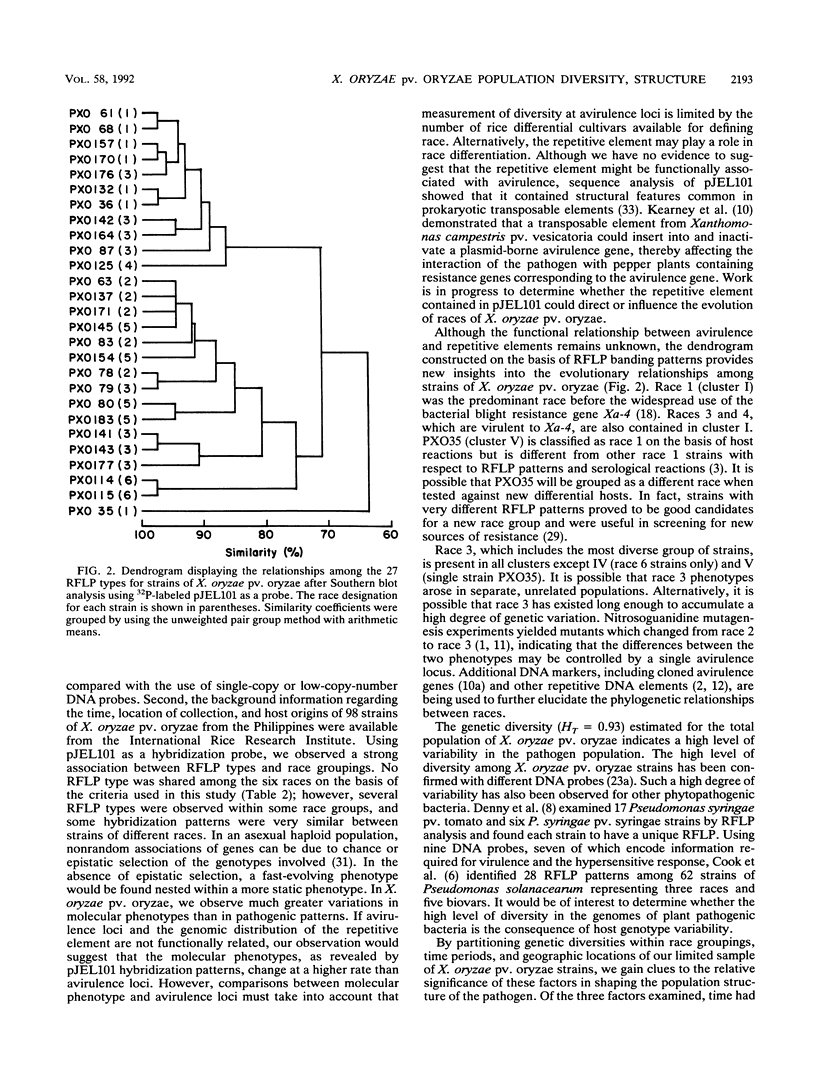
A repetitive DNA element cloned from Xanthomonas oryzae pv. oryzae was used to assess the population structure and genetic diversity of 98 strains of X. oryzae pv. oryzae collected between 1972 and 1988 from the Philippine Islands. Genomic DNA from X. oryzae pv. oryzae was digested with EcoRI and analyzed for restriction fragment length polymorphisms (RFLPs) with repetitive DNA element as a probe. Twenty-seven RFLP types were identified; there was no overlap of RFLP types among the six races from the Philippines. Most variability (20 RFLP types) was found in strains of races 1, 2, and 3, which were isolated from tropical lowland areas. Four RFLP types (all race 5) were found among strains isolated from cultivars grown in the temperate highlands. The genetic diversity of the total population of X. oryzae pv. oryzae was 0.93, of which 42% was due to genetic differentiation between races. The genetic diversities of strains collected in 1972 to 1976, 1977 to 1981, and 1982 to 1986, were 0.89, 0.90, and 0.92, respectively, suggesting a consistently high level of variability in the pathogen population over the past 15 years. Cluster analysis based on RFLP banding patterns showed five groupings at 85% similarity. The majority of strains from a given race were contained within one cluster, except for race 3 strains, which were distributed in three of the five clusters.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Denny T. P., Gilmour M. N., Selander R. K. Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J Gen Microbiol. 1988 Jul;134(7):1949–1960. doi: 10.1099/00221287-134-7-1949. [DOI] [PubMed] [Google Scholar]
- Nei M., Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics. 1981 Jan;97(1):145–163. doi: 10.1093/genetics/97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. J., Borman P. A rapid biochemical method for purifying high molecular weight bacterial chromosomal DNA for restriction enzyme analysis. Nucleic Acids Res. 1987 Apr 24;15(8):3631–3631. doi: 10.1093/nar/15.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Geographic components of linkage disequilibrium in natural populations of Escherichia coli. Mol Biol Evol. 1983 Dec;1(1):67–83. doi: 10.1093/oxfordjournals.molbev.a040302. [DOI] [PubMed] [Google Scholar]