Abstract
Maintenance of skeletal and cardiac muscle structure and function requires precise control of the synthesis, assembly, and turnover of contractile proteins of the sarcomere. Abnormalities in accumulation of sarcomere proteins are responsible for a variety of myopathies. However, the mechanisms that mediate turnover of these long-lived proteins remain poorly defined. We show that muscle RING finger 1 (MuRF1) and MuRF3 act as E3 ubiquitin ligases that cooperate with the E2 ubiquitin–conjugating enzymes UbcH5a, -b, and -c to mediate the degradation of β/slow myosin heavy chain (β/slow MHC) and MHCIIa via the ubiquitin proteasome system (UPS) in vivo and in vitro. Accordingly, mice deficient for MuRF1 and MuRF3 develop a skeletal muscle myopathy and hypertrophic cardiomyopathy characterized by subsarcolemmal MHC accumulation, myofiber fragmentation, and diminished muscle performance. These findings identify MuRF1 and MuRF3 as key E3 ubiquitin ligases for the UPS-dependent turnover of sarcomeric proteins and reveal a potential basis for myosin storage myopathies.
Introduction
Maintenance of striated muscle structure and function requires precise control of protein synthesis, processing, and degradation; abnormalities in these processes can give rise to myopathies (1). The ubiquitin proteasome system (UPS) is largely responsible for the degradation of misfolded proteins as well as long-lived proteins such as components of the contractile apparatus of striated muscles (2). Although activation of the UPS in skeletal muscle and heart has been primarily linked to a reduction in muscle mass as occurs during muscle atrophy (3, 4), recent evidence has also shown the UPS to be activated during cardiac hypertrophy (5). Little is known of the molecular basis of these apparently opposing functions of the UPS.
Substrate specificity of the UPS is mediated by 3 groups of E3 ubiquitin ligases, called RING finger, homologous to E6AP carboxyl terminus domain (HECT domain), and Skp1–Cul1–F-box protein complex (SCF complex) (6). Muscle RING finger (MuRF) proteins 1, 2, and 3 comprise a subfamily of the RING-finger E3 ubiquitin ligases that are expressed specifically in skeletal muscle and the heart (7). MuRF1 is upregulated during skeletal muscle atrophy, and mice lacking MuRF1 are resistant to atrophy (8). MuRF1 has also been implicated in cardiac hypertrophy (9). MuRF3 associates with microtubules and participates in the formation of cellular microtubular networks (7). Recently we reported that MuRF3 is involved in maintaining cardiac function and the integrity of the ventricular wall following acute myocardial infarction (10). Given the similarity in structure and expression of MuRF proteins, it is likely that the phenotypes of mutant mice lacking individual MuRF proteins may reveal only a subset of MuRF functions due to redundancy.
The specific interaction partners of MuRF proteins and the consequences of such interactions are only beginning to be determined. MuRF proteins have been shown to localize to the sarcomere (7, 11, 12) and are capable of forming heterodimers (11). MuRF1 associates with titin at the M-band of the sarcomere, which has been proposed to maintain stability of the sarcomeric M-line region (13, 14). MuRF1 was also reported to function as an E3 ubiquitin ligase that catalyzes the ubiquitination of troponin I (15) and creatine kinase (16). MuRF1 and MuRF2, but not MuRF3, interact with titin, nebulin, troponin T, myotilin, myosin light chain 2, and T-cap, whereas MuRF1, MuRF2, and MuRF3 interact with troponin I (17). Recently we showed that MuRF3 interacts with and mediates degradation of γ-filamin and four-and-a-half LIM domain 2 (10). It remains to be determined whether all of these MuRF interacting proteins serve as substrates for ubiquitination and UPS-dependent degradation or as scaffolding proteins.
Here we show that MuRF1–/–MuRF3–/– double mutant (DKO) mice display a distinct skeletal muscle and cardiac myopathy reminiscent of myosin storage myopathy (MSM) in humans. Skeletal and cardiac muscles of DKO mice display a striking subsarcolemmal accumulation of myosin heavy chain (MHC) accompanied by reduced maximal force development, cardiac hypertrophy, and decreased cardiac function. We demonstrate that MuRF1 and MuRF3 interact specifically with β/slow MHC and MHCIIa and utilize UbcH5a, -b, and -c as E2 ubiquitin–conjugating enzymes to catalyze the ubiquitination and degradation of these contractile proteins. We conclude that MuRF1 and MuRF3 play a central role in the maintenance of skeletal muscle and cardiac structure and function, at least in part, by regulating MHC ubiquitination and degradation.
Results
DKO mice display a skeletal muscle myopathy.
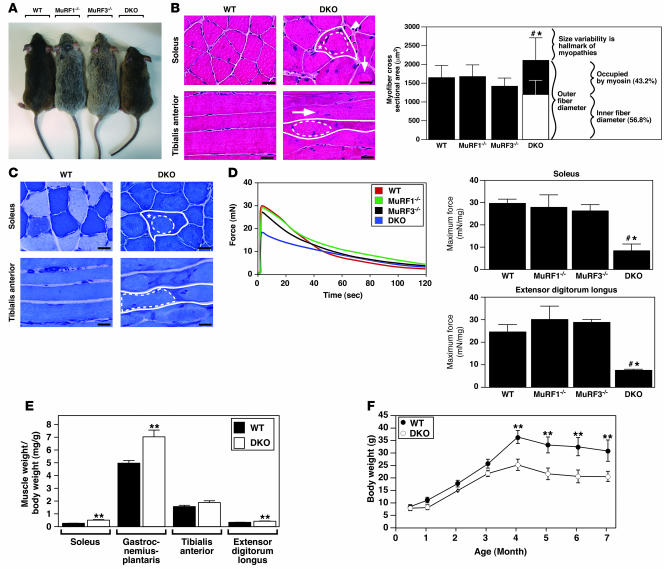
To determine the roles of MuRF1 and MuRF3 in vivo and to identify possible redundant targets of these putative E3 ubiquitin ligases, we generated DKO mice. Although no obvious phenotype was detected in MuRF1–/– (8, 9) or MuRF3–/– (10) mice, DKO mice were less mobile and displayed a myopathic gait, difficulty walking and climbing, and a reduced step length compared with WT and single mutant littermates (data not shown). This phenotype, affecting both male and female mice, included a significant decrease in body weight and size (Figure 1A) that was exacerbated with age (Figure 1F) and was accompanied by increased skeletal muscle mass (Figure 1E).
Figure 1. DKO mice show a skeletal muscle myopathy.
(A) Mice at 12 weeks of age. (B) H&E stain and (C) myosin ATPase activity of cross-sections from soleus and longitudinal sections from tibialis anterior muscles from 12-week-old WT and DKO mice. Subsarcolemmal eosinophilic material accumulated along the entire length of the myofiber around a central myofiber core of DKO muscle. Heterogeneity of fiber size, atrophic myofibers (arrowhead), and centrally localized nuclei (long arrow) were found in DKO muscle. Solid lines indicate outer myofiber membrane. Dotted lines define the central core, which is surrounded by accumulated material (asterisk). Short arrow indicates basophilic boundary surrounding inner fiber. Quantitation of myofiber cross-sectional area and determination of occupied surface area of myofibers from soleus muscle (n = 6 each). (D) Measurement of force development and fatigability. Representative recordings of force from soleus muscles. Quantitation of maximum force development from soleus and extensor digitorum longus (n = 6 each). mN, millinewtons. (E) Quantitation of skeletal muscle mass (n = 12 each) of 12-week-old mice. (F) Measurement of body weight with increasing age. Scale bars: 20 μm. #P < 0.01 versus WT and MuRF1–/–; *P < 0.01 versus MuRF3–/–; **P < 0.01 versus WT.
Histological analysis showed a myopathic phenotype in soleus, tibialis anterior (Figure 1B and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI32827DS1), extensor digitorum longus (Supplemental Figure 1), gastrocnemius-plantaris muscles, and diaphragm (data not shown) of DKO animals. Strikingly, the myopathy was characterized by the subsarcolemmal accumulation of protein along the entire length of the myofiber surrounding a central myofiber core. In addition, myofibers from DKO animals showed an abnormal variability in fiber size as well as atrophic and split myofibers, an increase in the number of nuclei per myofiber, and centralized nuclei indicative of regeneration (Figure 1B and Supplemental Figure 1). Fiber type analysis of DKO skeletal muscle using a metachromatic dye ATPase assay (18) revealed intense metachromatic staining around the perimeter and along the entire longitudinal lengths of DKO myofibers and showed no fiber type preference within the myopathic skeletal muscle (Figure 1C and Supplemental Figure 2). When soleus and extensor digitorum longus muscles were isolated and subjected to continuous electrical stimulation to measure muscle performance, we observed a profound reduction in maximum force in DKO mice compared with MuRF1–/–, MuRF3–/–, and WT littermates, with no significant changes in fatigability (Figure 1D).
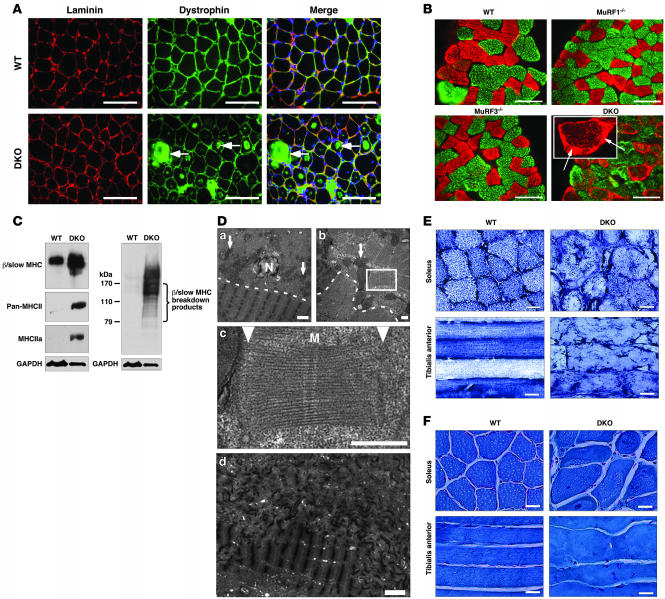
H&E staining of skeletal muscle from DKO mice revealed a basophilic demarcation separating the central myofiber core from the surrounding accumulating material (Figure 1B). Immunohistochemistry using anti-laminin and anti-dystrophin antibodies (Figure 2A) and electron microscopy (Figure 2D) showed no evidence of a limiting membrane separating the central myofiber core from the accumulating material, indicating that the accumulating material was contained within the DKO myofiber rather than an adjacent cell. Interestingly, some individual myofibers of the DKO soleus muscle were immunoreactive for dystrophin (Figure 2A), a condition reported as a general characteristic of human skeletal muscle myopathies (19).
Figure 2. DKO mice display MSM.
(A) Immunohistochemistry of soleus muscle using anti-laminin and anti-dystrophin antibodies. Dystrophin immunostaining was observed within myofibers only in DKO muscle. (B) Immunohistochemistry of soleus muscle using anti–β/slow MHC (red) and anti–pan-MHCII (green) antibodies. Inset shows a representative myofiber with subsarcolemmal β/slow MHC accumulation (arrows). (C) Immunoblotting of proteins from the soluble (left blot) and particulate (right blot) fractions of the soleus revealed an accumulation of β/slow MHC in DKO muscle. Anti-GAPDH was used as a control. (D) Electron microscopy of longitudinal (top left and bottom) and cross (top right and middle) sections of soleus muscle displayed accumulation of granular material and unassociated myosins in DKO myofibers. Arrows indicate mitochondria. Dotted lines indicate separation of inner myofiber core (below the line) from accumulating myosin (above the line). Middle panel shows an enlargement of the area within the box at top right. Arrowheads point toward abnormal Z-lines. Bottom: Z-line streaming found in skeletal muscle myopathies is seen in the top of this panel. M, M-line; N, nucleus. (E) NADH stain of soleus and tibialis anterior muscles shows disorganized mitochondria in DKO muscle. (F) Modified Gomori trichrome stain of soleus and tibialis anterior muscles shows no ragged red fiber myopathy. Scale bars: 100 μm (A and B); 2 μm (D, top left and bottom); 0.5 μm (D, top right and middle); 20 μm (E and F).
Abnormal accumulation of myosins in DKO myofibers.
Analysis of proteins isolated from soleus of WT, MuRF1–/–, MuRF3–/–, and DKO mice by SDS-PAGE revealed a band of approximately 60 kDa that was specific to DKO muscle (data not shown). Mass spectrometry identified this band as fragments of myosin proteins, which normally migrate at about 200 kDa (Supplemental Table 1). Immunohistochemistry of DKO soleus muscle using anti–β/slow MHC and anti–pan-MHCII antibodies confirmed the accumulation of both proteins within their respective myofibers (Figure 2B). MHC proteins accumulated as an amorphous homogenous mass surrounding a central myofiber core (Figure 2B). Immunoblot analysis of isolated protein from WT and DKO skeletal muscle showed a massive accumulation of undegraded and degraded β/slow MHC in DKO soleus (Figure 2C) and revealed that the increase in MHCII protein was mainly MHCIIa (Figure 2C) in DKO soleus.
Electron microscopy performed on sections of soleus showed protein aggregates only in DKO myofibers, leading to a redistribution of mitochondria along the centralized myofiber (Figure 2D). This was confirmed by NADH staining (Figure 2E and Supplemental Figure 3).
As observed in MSM (20, 21), we found that the DKO myofibers were degraded into granular material that accumulated around a central myofiber core (Figure 2D). The amorphous material contained unassociated filaments and fragments of sarcomeres. Measurements of the filaments in cross and transverse sections showed the filament thickness to be 15 nm, consistent with that of myosin filaments (22), validating the identity of the granular material as MHC protein (data not shown). Disoriented sarcomeres seen in cross and longitudinal sections of mutant DKO skeletal muscle had an intact M-line structure with adjacent thick filaments, suggesting a role of MuRF1 and MuRF3 in the turnover of thick filaments and M-line proteins. In contrast, the Z-line was greatly diminished or absent (Figure 2D), suggesting that mechanisms independent of MuRF1 and MuRF3 mediate degradation of Z-line proteins. In addition, disorganized and non-aligned, blurred, broadened, and disrupted Z-lines, known as Z-line streaming, a general feature of many myopathic disorders, was observed in skeletal muscle of DKO mice (Figure 2D).
To determine whether the myopathy seen in DKO muscle resembled nemaline myopathy, another striated muscle disorder, Gomori trichrome staining was performed. The results showed no evidence of ragged-red fibers, a hallmark of nemaline body myopathy (23) (Figure 2F and Supplemental Figure 4).
DKO mice exhibit hypertrophic cardiomyopathy.
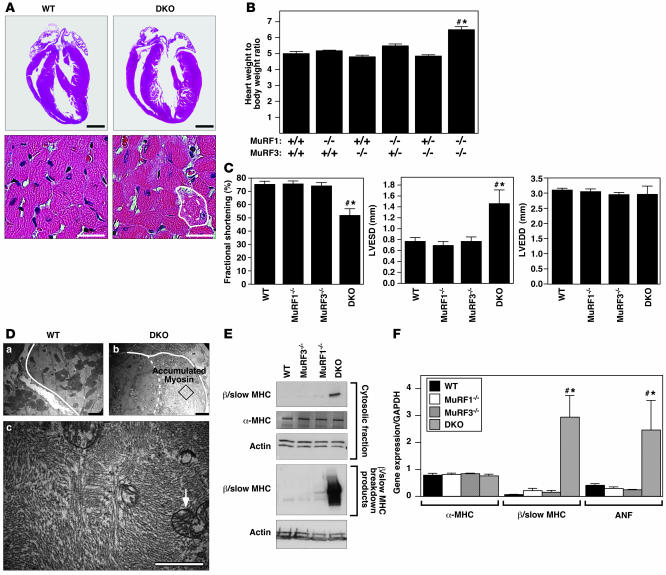
Hearts of DKO mice were hypertrophic, indicative of cardiac stress (Figure 3, A and B). Echocardiography showed a pronounced reduction of LV systolic function due primarily to an enlargement of the LV end-systolic dimension, whereas the LV end-diastolic dimension remained unchanged (Figure 3C). As in skeletal muscle, cardiomyocytes of DKO animals contained a sharply demarcated accumulation of subsarcolemmal protein composed of granular material and unassociated myosins. Mitochondria were redistributed along the centralized myofiber core (Figure 3, A and D). Immunoblotting showed an accumulation of β/slow MHC protein and its breakdown products in hearts of DKO compared with MuRF1–/–, MuRF3–/–, or WT littermates (Figure 3E). In contrast, expression of α-MHC, the adult cardiac myosin isoform, was not changed (Figure 3E). Atrial natriuretic factor (ANF), a marker of cardiac stress, was upregulated in DKO hearts compared with littermate controls (Figure 3F).
Figure 3. DKO mice exhibit hypertrophic cardiomyopathy and decreased cardiac function.
(A) H&E stain of hearts. Solid and dotted lines indicate outer myofiber and inner core diameter, respectively. (B) Heart weight/body weight ratios showed cardiac hypertrophy in DKO mice. (C) Echocardiography revealed decreased LV systolic function (fractional shortening), increased LV end-systolic diameter (LVESD), and an unchanged LV end-diastolic diameter (LVEDD). (D) Electron microscopy of heart showed accumulated and disoriented thick filaments and displacement of mitochondria (arrow). Solid and dotted lines indicate outer myofiber and inner core diameter, respectively. Bottom panel is magnification the box in the top right panel, showing ultrastructure of accumulated myosin filaments in the cardiomyocyte. (E) Immunoblotting of proteins from hearts revealed accumulation of undegraded (cytosolic fraction; top 3 panels) and degraded (pellet fraction; bottom 2 panels) β/slow MHC in DKO muscle. α-MHC expression remained unchanged. (F) Expression of hypertrophic markers ANF and β/slow MHC as assessed by real-time PCR analysis. α-MHC expression remained unchanged. #P < 0.01 versus WT and MuRF1–/–; *P < 0.01 versus MuRF3–/–. Scale bars: 1 mm (A, top); 20 μm (A, bottom); 2 μm (D).
MuRF1 and MuRF3 interact with and ubiquitinate MHC proteins in vitro.
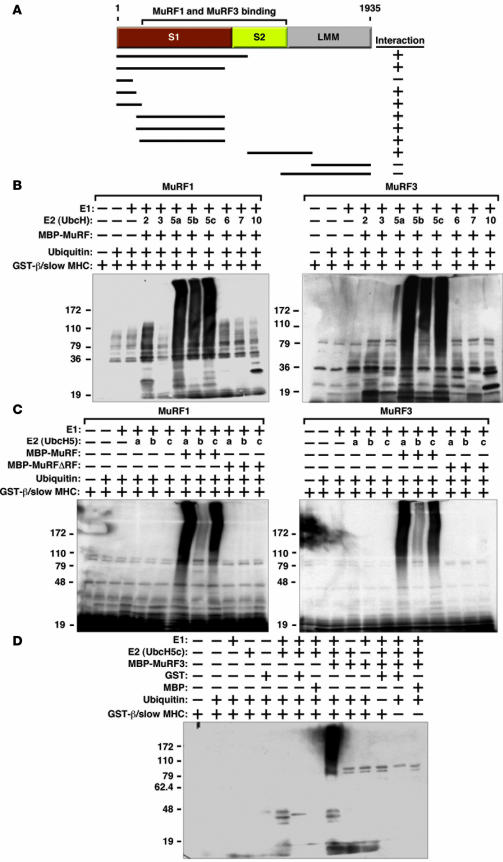
To begin to define the mechanistic basis of MHC accumulation in DKO muscles, we tested whether MuRF1 and MuRF3 interact with β/slow MHC and MHCIIa proteins. Indeed, the interaction of MuRF1 and MuRF3 proteins with the N-terminal regions of β/slow MHC and MHCIIa was readily apparent by coimmunoprecipitation assays using transfected COS-7 cells in vitro (Figure 4A). Binding was mediated by the S1 and S2 subfragments of myosin, which contain the globular head domain, responsible for ATP-dependent cross-bridge formation between thick and thin filaments, and a long α-helix domain, which is important for myosin dimerization. We used recombinant proteins encoding the N-terminal MuRF-interacting regions of β/slow MHC and MHCIIa protein for subsequent in vitro ubiquitination assays. We found that both MuRF1 and MuRF3 ubiquitinate residues within the N terminus of β/slow MHC (Figure 4B) as well as MHCIIa (Supplemental Figure 5A). We then systematically tested a series of E2 ubiquitin–conjugating enzymes for their ability to cooperate with MuRF1 and MuRF3 to ubiquitinate β/slow MHC and MHCIIa and found that MuRF1 and MuRF3 use the E2 ubiquitin–conjugating enzymes UbcH5a, -b, and -c and, to a lesser degree, UbcH2 to execute this reaction (Figure 4B and Supplemental Figure 5A). No differences in ubiquitination efficiency between these E2 ubiquitin–conjugating enzymes and MuRF1 and MuRF3 were observed, indicating that both MuRF proteins use the same E2 ubiquitin–conjugating enzymes for the ubiquitination of β/slow MHC and MHCIIa. In contrast, the E2 ubiquitin–conjugating enzymes UbcH3, -6, -7, and -10 did not facilitate ubiquitination by MuRF1 and MuRF3 (Figure 4B and Supplemental Figure 5A). RING-finger deletion mutants of MuRF1 and MuRF3 were unable to ubiquitinate β/slow MHC or MHCIIa regardless of the E2 ubiquitin–conjugating enzyme used (Figure 4C and Supplemental Figure 5B). In control experiments, no unspecific ubiquitination signal was detected, indicating specificity of the in vitro ubiquitination assays (Figure 4D).
Figure 4. MuRF1 and MuRF3 interact with and ubiquitinate β/slow MHC in vitro.
(A) Schematic diagram of interaction mapping between MuRF1 and MuRF3 and β/slow MHC shows that MuRF1 and MuRF3 interact with multiple domains of β/slow MHC. S1, subfragment 1; LMM, light meromyosin. (B) In vitro ubiquitination assays were performed with recombinant E1 ubiquitin–activating enzymes, E2 ubiquitin–conjugating enzymes, MBP-MuRF1 or MBP-MuRF3, ubiquitin, and GST–β/slow MHC. (B, C, and D) Ubiquitin protein conjugates were resolved by SDS-PAGE and detected by anti-ubiquitin antibody. Ubiquitination activity as assessed by the detection of high molecular weight multiubiquitin chains. (C) In vitro ubiquitination assays were performed with recombinant E1; UbcH5a, -5b, and -5c; ubiquitin; GST–β/slow MHC; MBP-MuRF1 and MBP-MuRF3; and RING-finger deletion mutants MBP-MuRF1ΔRF and MBP-MuRF3ΔRF. Ubiquitination activity of MuRF1 and MuRF3 toward GST–β/slow MHC was abolished when the RING-finger domains of MuRF1 and MuRF3 were deleted. (D) In vitro ubiquitination assays were performed with different combinations of recombinant E1, UbcH5c, ubiquitin, GST–β/slow MHC, and MBP-MuRF3. No ubiquitin protein conjugates were detected when GST or MBP proteins (2 far right lanes of panel) were used instead of GST–β/slow MHC, confirming the specificity of the assay.
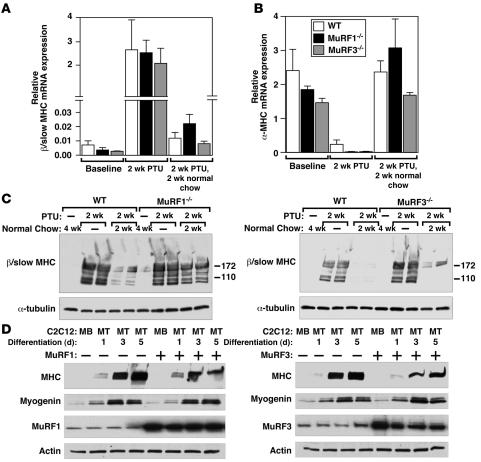
MuRF1 and MuRF3 mediate β/slow MHC ubiquitination and turnover in vivo.
β/Slow MHC is upregulated in the heart in response to hypothyroidism. To further examine the ability of MuRF1 and MuRF3 to mediate the ubiquitin-dependent turnover of β/slow MHC in vivo, we induced the expression of β/slow MHC in the heart by administering propylthiouracil (PTU), an inhibitor of T3 biosynthesis (24), to MuRF1–/–, MuRF3–/–, and WT mice for 2 weeks (Figure 5A). We used only the single MuRF mutant mice to induce β/slow MHC in the heart with PTU because the amount of β/slow MHC in the single MuRF mutant mouse is similar to that in WT mice (Figure 5A) and because DKO mice had a high baseline content of β/slow MHC in the heart (Figure 3E). Hypothyroidism led to a comparable increase in β/slow MHC (Figure 5A) and a decrease in α-MHC mRNA (Figure 5B) in the hearts of WT, MuRF1–/–, and MuRF3–/– mice. After withdrawal of PTU for 2 subsequent weeks, we compared the amount of β/slow MHC protein present as a readout for β/slow MHC degradation in the heart. Although β/slow MHC mRNA decreased in WT, MuRF1–/–, and MuRF3–/– hearts to similar levels, the content of β/slow MHC protein was elevated in the hearts of MuRF1–/– and MuRF3–/– mutant mice compared with WT littermates (Figure 5C). This finding suggests that the absence of either MuRF1 or 3 leads to a reduction of β/slow MHC degradation in the heart in vivo.
Figure 5. MuRF1 and MuRF3 mediate MHC protein turnover in vivo.
(A and B) PTU was administered for 2 weeks to induce expression of β/slow MHC in WT, MuRF1–/–, and MuRF3–/– mouse hearts (n = 6 each). Expression of β/slow MHC (A) and α-MHC (B) mRNA as assessed by real-time PCR analysis was increased and decreased, respectively, to comparable levels after 2 weeks of PTU treatment in all genotypes. Terminating the administration of PTU treatment led to a normalization of β/slow MHC and α-MHC mRNA expression. (C) Immunoblotting of proteins from hearts of WT, MuRF1–/–, and MuRF3–/– mice showed a reduced degradation of β/slow MHC in MuRF1–/– and MuRF3–/– compared with WT mice. Anti-tubulin antibody was used as a control. (D) Immunoblots of lysates from C2C12 myoblasts (MB) and differentiating myotubes (MT; days of differentiation are indicated at top) transfected with MuRF1-myc or MuRF3-myc, using anti-MHC antibody show a reduced MHC content when MuRF1 or MuRF3 were coexpressed. Transfection of pcDNA3.1 was used as a control. Anti-myogenin, was used as a control for differentiation, and anti-myc and anti-tubulin antibodies were used as controls for input.
MuRF1 and MuRF3 mediate increased myosin degradation in vitro.
To further examine the ability of MuRF1 and MuRF3 to promote myosin degradation, we overexpressed MuRF1 or MuRF3 in C2C12 myoblasts and analyzed MHC protein expression during myotube differentiation, which occurs when MHC proteins accumulate (25). We observed less accumulation of MHC in differentiating C2C12 myotubes in the presence of MuRF1 or MuRF3, suggesting that MuRF1 and MuRF3 inhibit accumulation of MHC in skeletal muscle cells (Figure 5D), whereas myogenin expression was similar in control cells and cells expressing MuRF protein, indicating that MuRF expression did not block myotube differentiation (Figure 5D).
Discussion
The results of this study show that MuRF1 and MuRF3 function redundantly to control the ubiquitin-dependent degradation of β/slow MHC and MHCIIa proteins in striated muscles. The deletion of both MuRF1 and MuRF3 in mice causes the abnormal accumulation of MHC thick filaments and ultrastructural abnormalities in skeletal and cardiac muscles, resulting in a severe decrease in muscle function. This phenotype and its consequences are reminiscent of the human myopathy MSM as well as hypertrophic cardiomyopathy, both of which have been linked to mutations within the MYH7 gene, which encodes β/slow MHC protein (26–33). However, in contrast to the human MSM, which is restricted to type I myofibers, we did not find a fiber type–restricted phenotype within DKO skeletal muscle. This difference from classical MSM, which is caused by a mutation the MYH7 gene encoding β/slow MHC (which is restricted to the heart and type I myofibers), reflects the lack of fiber type specificity of expression of MuRF1 and MuRF3 as well as their apparent ability to ubiquitinate MHCIIa and β/slow MHC. Thus a fiber type–restricted phenotype in DKO mice would not be expected. In addition, not all patients with MSM display a myopathy of both the skeletal muscle and the heart (26–33), whereas both muscles were affected in our DKO mice. Despite these differences we believe that the accumulation of myosin and the reduced performance of the affected muscle justifies the description of the phenotype of the DKO mouse as a MSM.
The mechanisms that mediate degradation of sarcomeric proteins have been the focus of intense interest (34). Loss of muscle proteins during cachexia (35), diabetes (36), sepsis (37), and neurological disorders (38) is thought to be mediated primarily by activation of the UPS. Although myosin proteins have been shown to be ubiquitinated (39), no specific E3 ubiquitin ligase or accompanying E2 ubiquitin–conjugating enzymes that mediate UPS-dependent MHC degradation have been identified. Moreover, although several interaction partners have been identified for MuRF3 (10), no E3 ubiquitin ligase activity has been demonstrated for MuRF3 to date. Here we show that β/slow MHC and MHCIIa serve as overlapping targets for ubiquitination and UPS-dependent degradation by MuRF1 and MuRF3 together with the E2 ubiquitin–conjugating enzymes UbcH5a, -b and -c.
Because MuRF proteins display both E3 ubiquitin ligase activity and play roles in stabilizing sarcomere structure and microtubules (7, 10, 11, 40), the phenotype of DKO mice might reflect not only the loss of ubiquitin ligase activity but also a loss of structural integrity of striated muscle cells. However, the accumulation of myosin in the absence of MuRF1 and MuRF3 and our data showing that MuRF1 and MuRF3 mediate ubiquitination and degradation of myosin strongly argue that the loss of MuRF1 and MuRF3 ubiquitin ligase activity contributes significantly to the phenotype of DKO mice.
Interestingly, although we found that MHC proteins are direct targets of MuRF1 and MuRF3, the absence of both E3 ubiquitin ligases did not affect the entire myofiber, as an apparently intact inner myofiber core was still present and surrounded by accumulating MHC proteins in DKO muscle. Thus it is tempting to speculate that MuRF1 and MuRF3 preferentially affect degradation of the soluble cytosolic fraction of MHC proteins rather than the MHC fraction that is associated with the sarcomere, which may actually be stabilized by MuRF proteins.
Because MuRF1 and MuRF3 promote the degradation of MHC proteins, we expected that loss of MuRF1 and MuRF3 would lead to accumulation of undegraded MHC. However, we observed an accumulation of both undegraded and degraded MHC in DKO muscle. This suggests that other MHC-degrading mechanisms, most likely non–proteasome dependent, may be activated in DKO muscle but are clearly not sufficient for complete MHC degradation. Several proteinases such as trypsin-like serine proteinase (41), calpain (42), cathepsin B/L (43, 44), and chymase (44) are activated during muscular dystrophy. In particular, trypsin-like serine proteinase mediates myosin cleavage (41) with myosin breakdown products similar to those we observed in DKO soleus (Figure 2C). Furthermore, treatment of myofibrils with a thiol proteinase resulted in the loss of the Z-disc (41), a finding analogous to our electron microscopy findings in which virtually no Z-disc structures were found within the accumulating MHC of DKO muscle. We therefore conclude that MuRF1 and MuRF3 are the key enzymes for UPS-dependent MHC degradation in striated muscle. This observation is important because the degradation of actomyosin complexes by the UPS is reduced compared with uncomplexed myosin and actin molecules (39) and the access of the sarcomere by the UPS is partially mediated by proteases such as caspase-3 (45) and calpain-1 (46), which are known to augment ubiquitination of structural proteins. It is therefore possible that activation of these proteases could be important for the initiation of myosin turnover in the myopathic DKO muscle and worsen the phenotype by producing substrates for UPS-dependent degradation. However, since MuRF1 and MuRF3 appear to be the rate-limiting enzymes regulating myosin ubiquitination and degradation, we believe that their absence is the primary cause of the DKO phenotype.
β/slow MHC is downregulated in the adult mouse heart but can be reversibly upregulated by pharmacological induction of hypothyroidism, providing a means of monitoring β/slow MHC degradation in the presence or absence of MuRF1 and MuRF3 proteins in vivo. We found that β/slow MHC degradation was greatly reduced in MuRF1 and MuRF3 single mutant mice compared with WT mice, further supporting a role of MuRF1 and MuRF3 in β/slow MHC degradation in the heart. Conversely, overexpression of MuRF1 and MuRF3 protein inhibited accumulation of MHC protein in differentiating C2C12 cells, indicating that MHC proteins are indeed targets for MuRF1- and MuRF3-dependent degradation. Interestingly, MuRF3 expression (7) parallels that of MHC (25) during C2C12 cell differentiation, which might reflect a mechanism by which myosin turnover is regulated during differentiation.
Nemaline myopathy, although clinically similar to myosin storage myopathies, is caused by mutations in a variety of genes encoding sarcomeric thin filament proteins, including nebulin (47), α-actin 1 (48), and troponin T1 (49). Although MuRF proteins have been shown to interact with nebulin and troponin T (17), our results using modified Gomori trichrome stain indicated that the phenotype observed in DKO muscle is mainly related to perturbed degradation and accumulation of myosin-thick but not -thin filaments. The accumulating MHC in DKO skeletal muscle caused a displacement of mitochondria, which likely resulted in inefficient energy supply to the contractile apparatus, thereby augmenting the phenotype in DKO muscle.
The cardiac phenotype observed in DKO mice consists of an increased heart weight, decreased cardiac function, and increased ANF expression. This phenotype is indicative of a hypertrophic cardiomyopathy and is likely to be caused by the accumulation of β/slow MHC protein in cardiomyocytes. Although we favor the hypothesis that the increase in β/slow MHC protein is mainly due to a lack of MuRF1- and MuRF3-dependent degradation, we are aware that β/slow MHC protein expression is also upregulated in response to cardiac stress. It is possible that the cardiac hypertrophy in DKO mice contributes to the accumulation of β/slow MHC observed in DKO hearts, further exacerbating the cardiac phenotype of DKO mice.
Muscular atrophy accompanies various clinically important diseases and determines the quality of life and mortality of patients. The development of novel therapeutic strategies to treat this disease requires a detailed understanding of the specific mechanisms that lead to the degradation of sarcomeric proteins. Indeed, MuRF1 plays a role in muscular atrophy (8), and it was proposed that MuRF inhibitors could be beneficial to treat skeletal muscle atrophy. However, based on our findings presented here and published earlier (10), administration of nonspecific MuRF inhibitors seems likely to cause adverse side effects, especially due to myosin accumulation in striated muscle cells. To be therapeutically effective, we propose that MuRF inhibitors should be designed to be isoform specific and to specifically disrupt the interaction between MuRF proteins and their targets.
Methods
Animal studies.
MuRF1 (8) and MuRF3 (10) mutant mice have been previously described. Animals were housed under standard conditions and maintained on commercial mouse chow and water ad libitum unless otherwise indicated. The environment was maintained at 22°C with a 12-hour light/12-hour dark cycle. MuRF1 and MuRF3 double heterozygous mutant mice were interbred, and resulting WT, MuRF1–/–, MuRF3–/–, and DKO littermates of the same gender were used for subsequent experiments. All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committees at the University of Texas Southwestern Medical Center.
Induction of hypothyroidism by administering PTU.
Hypothyroidism of 8-week-old male WT, MuRF1–/–, and MuRF3–/– mice (n = 6 each) was induced by feeding PTU, a commonly used anti-thyroid drug (24). Animals were divided into 2 groups; group 1 was sacrificed after 2 weeks of PTU treatment (n = 3 each), and the PTU diet for group 2 was changed to normal chow after 2 weeks of PTU treatment for an additional 2 weeks (n = 3 each). PTU-treated mice received 0.15% PTU feed (Harlan Teklad). WT, MuRF1–/–, and MuRF3–/– mice that received regular feed and drinking water for 4 weeks served as controls (n = 3 each). Hearts were flash-frozen in liquid nitrogen and stored at –80°C for subsequent analysis.
Muscle fatigue measurements.
Measurements of muscle fatigue were performed as previously described (50). To compare muscle function of WT, MuRF1–/–, MuRF3–/–, and DKO mice, male and female littermates and mice of the same age were analyzed. The soleus and extensor digitorum longus muscles were isolated from anesthetized mice and immediately mounted between a fixed clamp at the base of a water-jacketed organ bath and a Grass FTO3C isometric force transducer. Muscles were equilibrated for 15 minutes in an oxygenated (95% O2 and 5% CO2) physiological salt solution containing 120.5 mM NaCl, 4.8 mM KCl, 1.2 mM MgSO4, 1.5 mM CaCl2, 1.2 mM Na2PO4, 20.4 mM NaHCO3, 10 mM dextrose, and 1 mM pyruvate, at 30°C and pH 7.6. The muscles were adjusted to the optimal length followed by 30 minutes of resting. Muscles were then stimulated at 100 Hz until the force output dropped to 10% of the initial level. The time taken to reach 10%, 30%, and 50% of the original force was measured and used as an indication of muscle fatigability. The force development in millinewtons was normalized to the muscle mass in milligrams.
Histology and electron microscopy analyses.
Skeletal muscle and hearts were harvested, fixed with 3.7% paraformaldehyde, processed, and stained with H&E as previously described (10). Images were acquired with a Leica DM2000 microscope and an Optronics DEI-750 charge-coupled device digital camera and analyzed using ImageJ software 1.62c (http://rsb.info.nih.gov/ij/) to calculate myofiber cross sectional area.
Electron microscopy was performed as previously described (10). Briefly, soleus muscle and tissue of the interventricular septum of the heart of 8-week-old male littermate WT, MuRF1–/–, MuRF3–/–, and DKO mice were dissected and fixed in 2% glutaraldehyde in 0.1 M cacodylate buffer overnight at 4°C. The tissue was postfixed and stained with 1% osmium tetroxide in 0.1 M cacodylate buffer, incubated with uranyl acetate, and dehydrated with ethanol. The tissues were subsequently rinsed with propylene oxide and embedded using EMBED 812. Longitudinal and thin cross-sections (60 nm) were counterstained with uranyl acetate and lead citrate and examined on a Jeol 1200 electron microscope. Photographs were taken at the magnifications indicated in the figure legends.
Immunohistochemical analysis and metachromatic assays of skeletal muscle.
Soleus, tibialis anterior, extensor digitorum longus, and gastrocnemius-plantaris muscles were harvested from WT, MuRF1–/–, MuRF3–/–, and DKO mice and either flash frozen in embedding medium containing a 3:1 mixture of Tissue Freezing Medium (Triangle Biomedical Sciences) and gum tragacanth (Sigma-Aldrich) or fixed in 4% paraformaldehyde and processed for routine paraffin histology. Frozen sections were cut on a cryotome and stained histochemically for ATPase enzyme activity or modified Gomori trichrome as previously described (18, 21); β-nicotinamide adenine dinucleotide reduced disodium salt hydrate and Gomori 1 Step Trichrome with Light Green were purchased from Sigma-Aldrich and Poly Scientific, respectively. Paraffin and frozen histological sections were stained immunohistochemically for β/slow MHC (NOQ7, monoclonal; Sigma-Aldrich), pan-MHCII (My32, monoclonal; Sigma-Aldrich), laminin (polyclonal; Sigma-Aldrich), and dystrophin (monoclonal; Chemicon).
Cell culture, transfection, and coimmunoprecipitation.
Cell culture experiments were performed using COS-7 and C2C12 cells maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin/streptomycin and transfected using FuGENE 6 Transfection Reagent (Roche Applied Science) and Lipofectamine and PLUS Reagent (Invitrogen) according to the manufacturer’s instructions, respectively. Differentiation of C2C12 cells was induced 24 hours after transfection for time points indicated in Figure 5D using DMEM supplemented with 2% horse serum, 2 mM l-glutamine, and penicillin/streptomycin. All experiments were conducted in duplicate and repeated at least 3 times.
Expression plasmids containing aminoterminal FLAG- and Myc-tagged derivatives of MuRF3 were previously published (7, 10). Expression plasmids containing aminoterminal FLAG-tagged and carboxyterminal Myc-tagged derivatives of MuRF1 and MuRF3 and aminoterminal Myc-tagged derivatives of β/slow MHC and MHCIIa were generated using HighFidelity Taq DNA polymerase using mouse cDNA as template. For recombinant proteins, full-length and RING-finger–deleted (∆RF) MuRF1 and MuRF3 were subcloned into pMAL-c2x expression plasmids (NEB), and DNA fragments encoding amino acids 2–128 of WT β/slow MHC and amino acids 2–255 WT MHCIIa were subcloned into the pGEX-4T1 vector.
For immunoprecipitation studies, cells were harvested in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF) and supplemented with protease inhibitors (Complete; Roche Diagnostics), and FLAG- and myc-tagged proteins were immunoprecipitated with M2-agarose conjugate (Sigma-Aldrich) and anti-myc antibody (polyclonal; Santa Cruz Biotechnology Inc.), respectively, as previously described (10). Bound proteins were resolved by SDS-PAGE, transferred to PVDF membranes, and immunoblotted using either anti-Myc (polyclonal, A-14; Santa Cruz Biotechnology Inc.) or anti-FLAG (monoclonal, M2; Sigma-Aldrich) antibody. Loading controls were detected with anti–α-tubulin (monoclonal; Sigma-Aldrich) or anti-GAPDH antibody (monoclonal; Millipore). Proteins were visualized with a chemiluminescence system (Santa Cruz Biotechnology Inc.). Western blot analysis was also performed on protein samples from mouse hearts and skeletal muscles, as previously described (10, 51) using anti–β/slow MHC (NOQ7, monoclonal; Sigma-Aldrich), anti–pan-MHCII (My32, monoclonal; Sigma-Aldrich), or anti-MHCIIa antibody (sc-71 antibody was a gift from Zhen Yan, Duke University, Durham, North Carolina, USA). Silver staining of proteins from cardiac muscle was performed as previously described (50). Loading controls were detected with anti-GAPDH (monoclonal; Millipore) and anti-actin (monoclonal; Sigma-Aldrich) antibody.
In vitro ubiquitination assay and mass spectrometry.
In vitro ubiquitination assays were performed as published (52). Briefly, 500 ng of MBP-MuRF1, MBP-MuRF1∆RF, MBP-MuRF3, or MBP-MuRF3∆RF; 500 ng of GST–β/slow MHC or GST-MHCIIa; 0.3 μM purified E1 ubiquitin–activating enzyme (Boston Biochem); 40 mg/ml ubiquitin (Sigma-Aldrich); 8 mM ATP (Sigma-Aldrich); and 1 μM of E2 ubiquitin–conjugating enzyme as indicated in Figure 4B (Boston Biochem) were incubated in 100 mM Tris-Cl, pH 7.6, 10 mM MgCl2, 1 mM DTT, 50 μM MG132, and 2 mM NaVO4 protease inhibitors (Complete; Roche Diagnostics) for 2 hours at 30°C. Ubiquitin-protein conjugates were resolved by SDS-PAGE and detected by anti-ubiquitin antibody (polyclonal; Santa Cruz Biotechnology Inc.). Ubiquitination activity was assessed by the detection of high-molecular-weight multiubiquitin chains. Mass spectrometry was performed as previously published (53).
Semiquantitative RT-PCR.
Total RNA was isolated from skeletal muscles and the left ventricle of mice using TRI zol Reagent (Invitrogen). Total RNA was DNAse digested, and cDNA synthesis of 1.5 μg of RNA was carried out using a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). Real-time PCR was performed using either SYBR Green (ABI) with primer sets and PCR conditions that have been previously published (50) or TaqMan Universal PCR Mastermix (Applied Biosystems) with commercially available primer and probe sets as previously described (10, 51).
Statistics.
Values are presented as mean ± SEM. Gene expression was normalized to GAPDH mRNA and calculated as relative change. Differences in morphologic, physiologic, and biochemical parameters between groups were analyzed by Mann-Whitney U test or 2-sided Student’s t test. Statistics were calculated with Microsoft Excel 2002 and SPSS 11.0 software. A P value of less than 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Dennis Burns for insightful comments and advice with the electron microscopy images, Laurie Muller for technical help with electron microscopy, Regeneron Pharmaceuticals for MURF1–/– mice, Jennifer Brown for editorial assistance, and Alisha Tizenor for graphics. We thank Igor Rybkin, Kunhua Song, and Matthew Potthoff for helpful discussions. E.N. Olson is supported by grants from the NIH, the Donald W. Reynolds Cardiovascular Clinical Research Center, and the Robert A. Welch Foundation. J. Fielitz was supported by a fellowship from the Muscular Dystrophy Association and the Pfizer fellowship of the German Society of Cardiology.
Footnotes
Nonstandard abbreviations used: ANF, atrial natriuretic factor; DKO, MuRF1–/–MuRF3–/–; MHC, myosin heavy chain; MSM, myosin storage myopathy; MuRF, muscle RING finger; PTU, propylthiouracil; UPS, ubiquitin proteasome system.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2486–2495 (2007). doi:10.1172/JCI32827
References
- 1.Wang X., Robbins J. Heart failure and protein quality control. Circ. Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. The ubiquitin proteolytic system: from a vague idea, through basic mechanisms, and onto human diseases and drug targeting. Neurology. 2006;66:S7–S19. doi: 10.1212/01.wnl.0000192261.02023.b8. [DOI] [PubMed] [Google Scholar]
- 3.Glass D.J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 4.Razeghi P., et al. Atrophic remodeling of the heart in vivo simultaneously activates pathways of protein synthesis and degradation. Circulation. 2003;108:2536–2541. doi: 10.1161/01.CIR.0000096481.45105.13. [DOI] [PubMed] [Google Scholar]
- 5.Depre C., et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 6.Powell S.R. The ubiquitin-proteasome system in cardiac physiology and pathology. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H1–H19. doi: 10.1152/ajpheart.00062.2006. [DOI] [PubMed] [Google Scholar]
- 7.Spencer J.A., Eliazer S., Ilaria R.L., Jr., Richardson J.A., Olson E.N. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J. Cell Biol. 2000;150:771–784. doi: 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodine S.C., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 9.Willis M.S., et al. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ. Res. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fielitz J., et al. Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4377–4382. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centner T., et al. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J. Mol. Biol. 2001;306:717–726. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 12.McElhinny A.S., Perry C.N., Witt C.C., Labeit S., Gregorio C.C. Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J. Cell Sci. 2004;117:3175–3188. doi: 10.1242/jcs.01158. [DOI] [PubMed] [Google Scholar]
- 13.Gotthardt M., et al. Conditional expression of mutant M-line titins results in cardiomyopathy with altered sarcomere structure. J. Biol. Chem. 2003;278:6059–6065. doi: 10.1074/jbc.M211723200. [DOI] [PubMed] [Google Scholar]
- 14.McElhinny A.S., Kakinuma K., Sorimachi H., Labeit S., Gregorio C.C. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J. Cell Biol. 2002;157:125–136. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedar V., et al. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao T.J., Yan Y.B., Liu Y., Zhou H.M. The generation of the oxidized form of creatine kinase is a negative regulation on muscle creatine kinase. J. Biol. Chem. 2007;282:12022–12029. doi: 10.1074/jbc.M610363200. [DOI] [PubMed] [Google Scholar]
- 17.Witt S.H., Granzier H., Witt C.C., Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J. Mol. Biol. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Frey N., et al. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 2004;10:1336–1343. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 19.Selcen D., Ohno K., Engel A.G. Myofibrillar myopathy: clinical, morphological and genetic studies in 63 patients. Brain. 2004;127:439–451. doi: 10.1093/brain/awh052. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez C., et al. Electron microscopy in neuromuscular disorders. Ultrastruct. Pathol. 2005;29:437–450. doi: 10.1080/01913120500323175. [DOI] [PubMed] [Google Scholar]
- 21.Tajsharghi H., et al. Myosin storage myopathy associated with a heterozygous missense mutation in MYH7. Ann. Neurol. 2003;54:494–500. doi: 10.1002/ana.10693. [DOI] [PubMed] [Google Scholar]
- 22.Ip W., Heuser J. Direct visualization of the myosin crossbridge helices on relaxed rabbit psoas thick filaments. J. Mol. Biol. 1983;171:105–109. doi: 10.1016/s0022-2836(83)80317-9. [DOI] [PubMed] [Google Scholar]
- 23.Schnell C., Kan A., North K.N. ‘An artefact gone awry’: identification of the first case of nemaline myopathy by Dr R.D.K. Reye. Neuromuscul. Disord. 2000;10:307–312. doi: 10.1016/s0960-8966(99)00123-6. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y., Peng J., Campbell K.B., Labeit S., Granzier H. Hypothyroidism leads to increased collagen-based stiffness and re-expression of large cardiac titin isoforms with high compliance. J. Mol. Cell. Cardiol. 2007;42:186–195. doi: 10.1016/j.yjmcc.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 25.Ono Y., Sensui H., Sakamoto Y., Nagatomi R. Knockdown of hypoxia-inducible factor-1alpha by siRNA inhibits C2C12 myoblast differentiation. J. Cell. Biochem. 2006;98:642–649. doi: 10.1002/jcb.20804. [DOI] [PubMed] [Google Scholar]
- 26.Barohn R.J., Brumback R.A., Mendell J.R. Hyaline body myopathy. Neuromuscul. Disord. 1994;4:257–262. doi: 10.1016/0960-8966(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 27.Bohlega S., et al. Autosomal dominant hyaline body myopathy: clinical variability and pathologic findings. Neurology. 2003;61:1519–1523. doi: 10.1212/01.wnl.0000096022.09887.9d. [DOI] [PubMed] [Google Scholar]
- 28.Dye D.E., Azzarelli B., Goebel H.H., Laing N.G. Novel slow-skeletal myosin (MYH7) mutation in the original myosin storage myopathy kindred. Neuromuscul. Disord. 2006;16:357–360. doi: 10.1016/j.nmd.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Laing N.G., et al. Myosin storage myopathy: slow skeletal myosin (MYH7) mutation in two isolated cases. Neurology. 2005;64:527–529. doi: 10.1212/01.WNL.0000150581.37514.30. [DOI] [PubMed] [Google Scholar]
- 30.Masuzugawa S., et al. Autosomal dominant hyaline body myopathy presenting as scapuloperoneal syndrome: clinical features and muscle pathology. Neurology. 1997;48:253–257. doi: 10.1212/wnl.48.1.253. [DOI] [PubMed] [Google Scholar]
- 31.Meredith C., et al. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1). Am. J. Hum. Genet. 2004;75:703–708. doi: 10.1086/424760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldfors A., Tajsharghi H., Darin N., Lindberg C. Myopathies associated with myosin heavy chain mutations. Acta Myol. 2004;23:90–96. [PubMed] [Google Scholar]
- 33.Shingde M.V., et al. Myosin storage (hyaline body) myopathy: a case report. Neuromuscul. Disord. 2006;16:882–886. doi: 10.1016/j.nmd.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Ventadour S., Attaix D. Mechanisms of skeletal muscle atrophy. Curr. Opin. Rheumatol. 2006;18:631–635. doi: 10.1097/01.bor.0000245731.25383.de. [DOI] [PubMed] [Google Scholar]
- 35.Acharyya S., et al. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J. Clin. Invest. 2004;114:370–378. doi: 10.1172/JCI200420174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lecker S.H., et al. Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J. Clin. Invest. 1999;104:1411–1420. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiao G., et al. Sepsis is associated with increased mRNAs of the ubiquitin-proteasome proteolytic pathway in human skeletal muscle. J. Clin. Invest. 1997;99:163–168. doi: 10.1172/JCI119143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wing S.S., Haas A.L., Goldberg A.L. Increase in ubiquitin-protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and atrophy denervation. Biochem. J. 1995;307:639–645. doi: 10.1042/bj3070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solomon V., Goldberg A.L. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J. Biol. Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 40.Gregorio C.C., Perry C.N., McElhinny A.S. Functional properties of the titin/connectin-associated proteins, the muscle-specific RING finger proteins (MURFs), in striated muscle. J. Muscle Res. Cell Motil. 2005;26:389–400. doi: 10.1007/s10974-005-9021-x. [DOI] [PubMed] [Google Scholar]
- 41.Kay J., Siemankowski R.F., Siemankowski L.M., Goll D.E. Degradation of smooth-muscle myosin by trypsin-like serine proteinases. Biochem. J. 1982;201:267–278. doi: 10.1042/bj2010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spencer M.J., Croall D.E., Tidball J.G. Calpains are activated in necrotic fibers from mdx dystrophic mice. J. Biol. Chem. 1995;270:10909–10914. doi: 10.1074/jbc.270.18.10909. [DOI] [PubMed] [Google Scholar]
- 43.Kominami E., Bando Y., Ii K., Hizawa K., Katunuma N. Increases in cathepsins B and L and thiol proteinase inhibitor in muscle of dystrophic hamsters. Their localization in invading phagocytes. J. Biochem. (Tokyo). 1984;96:1841–1848. doi: 10.1093/oxfordjournals.jbchem.a135018. [DOI] [PubMed] [Google Scholar]
- 44.Tseng B.S., et al. Regenerated mdx mouse skeletal muscle shows differential mRNA expression. J. Appl. Physiol. 2002;93:537–545. doi: 10.1152/japplphysiol.00202.2002. [DOI] [PubMed] [Google Scholar]
- 45.Du J., et al. Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J. Clin. Invest. 2004;113:115–123. doi: 10.1172/JCI200418330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galvez A.S., et al. Cardiomyocyte degeneration with calpain deficiency reveals a critical role in protein homeostasis. Circ. Res. 2007;100:1071–1078. doi: 10.1161/01.RES.0000261938.28365.11. [DOI] [PubMed] [Google Scholar]
- 47.Wallgren-Pettersson C., et al. Mutations in the nebulin gene can cause severe congenital nemaline myopathy. Neuromuscul. Disord. 2002;12:674–679. doi: 10.1016/s0960-8966(02)00065-2. [DOI] [PubMed] [Google Scholar]
- 48.Goebel H.H., Anderson J.R., Hubner C., Oexle K., Warlo I. Congenital myopathy with excess of thin myofilaments. Neuromuscul. Disord. 1997;7:160–168. doi: 10.1016/s0960-8966(97)00441-0. [DOI] [PubMed] [Google Scholar]
- 49.Johnston J.J., et al. A novel nemaline myopathy in the Amish caused by a mutation in troponin T1. Am. J. Hum. Genet. 2000;67:814–821. doi: 10.1086/303089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh M., et al. Calcineurin is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol. Cell. Biol. 2005;25:6629–6638. doi: 10.1128/MCB.25.15.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bush E., et al. A small molecular activator of cardiac hypertrophy uncovered in a chemical screen for modifiers of the calcineurin signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2870–2875. doi: 10.1073/pnas.0308723101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ea C.K., Sun L., Inoue J., Chen Z.J. TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15318–15323. doi: 10.1073/pnas.0404132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhi G., et al. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17519–17524. doi: 10.1073/pnas.0506846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.