Abstract
Procedures to diagnose renal allograft rejection depend upon detection of graft dysfunction and the presence of a mononuclear leukocytic infiltrate; however, the presence of a modest cellular infiltrate is often not conclusive and can be detected in non-rejecting grafts. We have pursued a molecular approach utilizing reverse transcription (RT)-PCR to test the diagnostic accuracy of multiple immune activation gene analysis as means to diagnose renal allograft rejection. The magnitude of intragraft gene expression of 15 immune activation genes was quantified by competitive RT-PCR in 60 renal allograft core biopsies obtained for surveillance or to diagnose the etiology of graft dysfunction. Results were compared with a clinicopathological analysis based upon the histological diagnosis (Banff criteria) and the response to antirejection treatment. During acute renal allograft rejection intragraft expression of the interleukin (IL)-7 (P < 0.001), IL-10 (P < 0.0001), IL-15 (P < 0.0001), Fas ligand (P < 0.0001), perforin (P < 0.0001), and granzyme B (P < 0.0015), but not IL-2, interferon γ, or IL-4, genes is significantly heightened. Amplified RANTES and IL-8 gene transcripts are sensitive but nonspecific markers of rejection. A simultaneous RT-PCR evaluation of perforin, granzyme B, and Fas ligand identifies acute rejection, including cases with mild infiltration, with extraordinary sensitivity (100%) and specificity (100%). Effective antirejection therapy results in a rapid down-regulation of gene expression. The combined analysis of Fas ligand, perforin, and granzyme B gene expression by quantitative RT-PCR provides a reliable tool for diagnosis and follow-up of acute renal allograft rejection. Its accuracy and a potential rapid application within few hours suggest its use in the clinical management of renal transplant patients.
Keywords: renal transplantation, cytokines, cytotoxic T lymphocyte, polymerase chain reaction, diagnosis
Despite the recent improvement in renal allograft survival, the loss of graft function due to acute and chronic rejection continues and is a leading cause of end-stage renal failure today. As the occurrence of acute rejection episodes is the most powerful predictive factor for the later development of chronic rejection in adults (1) and children (2), many advocate strategies to detect and ablate acute rejection episodes as early as possible.
Antigen-triggered T-cell activation and the subsequent infiltration of activated CD4+ and CD8+ T-cell clones, macrophages, and natural killer (NK) cells into the graft are key events of acute allograft rejection. Although a T-cell-rich interstitial nephritis is a hallmark of acute allograft rejection, clinical rejection episodes responsive to treatment not infrequently show only a modest cellular infiltrate (borderline cases) (3), and similar infiltrates have been observed in surveillance biopsies obtained in well-functioning renal allografts (4, 5).
T-cell activation is characterized in large measure by a preprogrammed sequence of tightly regulated gene expression events in which genes are activated and silenced in their preordained order (6). Insofar as allograft rejection is a T-cell-dependent process, we hypothesized that clinical rejection is associated with expression of a specific subset of T-cell-dependent immune activation genes that may serve as a diagnostic indicator of rejection. We have hypothesized that patterns of intragraft mRNA generation during a cytopathic allograft response will be substantially different from those seen in other causes of graft dysfunction and may provide timely and specific information of immune events relevant to graft rejection.
This idea is supported by data derived from both animal models and clinical transplantation. In a mouse model of islet allograft rejection interleukin (IL)-2 and interferon (IFN)-γ gene expression preceded but did not accompany the onset of hyperglycemia, while transcripts for the cytotoxic T-lymphocyte (CTL)-selective granzyme B (GB) gene were observed during graft rejection (7). In clinical renal allograft rejection IL-2 transcripts are only rarely observed, while expression of IL-10 (8) and IL-15 genes (9) accompanies apparent rejection episodes. The presence of activated CTLs (10, 11) and expression of Ca2+-dependent perforin (P) and/or GB have been described in acute rejection of human hearts (12–14), lungs, (15) and kidneys (16–18). Unlike the situation in frank rejection, T-cell infiltrates in well-functioning grafts do not express cytokines, GB, or P (19).
The more recently discovered Fas ligand (FasL)/Fas receptor-mediated CTL injury initiates target cell death via a Ca2+-independent apoptotic pathway. Intragraft FasL expression, noted during murine cardiac allograft rejection (20), has not been investigated in clinical transplantation.
The overall aim of our study was to utilize competitive reverse transcription (RT)-PCR as a highly sensitive approach to evaluate the pattern of immune activation gene expression specific for rejection and to test the reliability of gene expression analysis as a diagnostic tool in kidney allograft rejection. In consideration of our previous experience, which linked expression of the CTL-specific P and GB genes to rejection, we sought to determine which immune activation genes regularly accompany allograft rejection, whether evaluation of both Ca2+-dependent and Ca2+-independent CTL pathways could strengthen the diagnostic accuracy of this approach, and, by evaluating sequential specimens, determine which immune activation genes may precede and, in the future, help to anticipate rejection episodes or predict failure of antirejection therapy.
MATERIALS AND METHODS
Biopsies.
Sixty kidney transplant biopsies were investigated for gene expression of chemokines [IL-8, RANTES (regulated upon activation, normal T-cell expressed and secreted)], T-cell growth factors and other cytokines (IL-2, IL-4, IL-7, IL-10, IL-15, and IL-17), cell surface immunoregulatory proteins (CTLA4), cytotoxic effector molecules (P, GB, FasL), IFN-γ, transforming growth factor (TGF)-β1, and the housekeeping protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Thirty-eight biopsies were obtained from 34 patients (25 adults and 9 children) to clarify the cause of graft dysfunction, 20 for early post-transplant surveillance and 2 from living related donor kidneys prior to reperfusion. Small portions of biopsy cores ((null)/1;10–½) were immediately snap frozen in liquid nitrogen at the bedside and stored at −70°C. The majority of tissue was used for histopathological analysis. Biopsies obtained to evaluate the cause of graft dysfunction were classified according to the Banff criteria (3) as rejection (pretreatment n = 12, post-treatment n = 3), nonrejection (acute tubular necrosis, cyclosporine nephrotoxicity n = 12), chronic rejection (n = 3), recurrence of primary disease (n = 4), or other complications (n = 4). In 4 of 12 rejecting samples and 4 of 12 acute tubular necrosis samples a mild cellular infiltrate was observed (borderline cases) and the diagnosis of rejection was confirmed by a beneficial clinical response to corticosteroids or OKT3 treatment.
RNA Isolation.
Procedures for isolation of tissue RNA and reverse transcription into cDNA were performed as previously described in detail (16). In brief, total RNA was isolated by tissue homogenization in guanidine isothiocyanate/2-mercaptoethanol and ultracentrifugation in CsCl. One microgram of RNA was reverse transcribed by Moloney murine leukemia virus transcriptase and diluted to a final volume of 40 μl.
Quantification of Gene Expression by Competitive Template RT-PCR.
Expression of specific gene transcripts identified within biopsy tissue was quantified by competitive RT-PCR. The cDNA derived from biopsy samples is coamplified with a known amount of a mutated target gene cDNA fragment—the gene-specific competitor. Sense and antisense oligonucleotides proportionately amplify both competitor and reverse-transcribed cDNA sequences in accordance with their relative initial abundance in the PCR. The PCR products are separated by agarose gel electrophoresis, stained with ethidium bromide, photographed in UV light with Polaroid type 55 positive/negative film, and scanned by laser densitometry (LKB Ultrascan). The ratio of densities (competitive template/reverse-transcribed cDNA) reflects the initial amounts of cDNA added (pg of competitive template per pg of reverse-transcribed cDNA). Standard curves were generated by serial dilutions of the gene-specific competitors with a constant amount of control reverse transcribed cDNA, thereby enabling quantification of the wild-type gene transcript.
Contaminating genomic DNA can be easily identified by size differences, as all oligonucleotide probes were targeted to separate exons of the gene of interest. The conditions used for all competitive PCRs were identical: 94°C for 30 sec, 55°C for 20 sec, 72°C for 20 sec, 10-min extension at 72°C after 35 cycles (Perkin–Elmer Cetus 480).
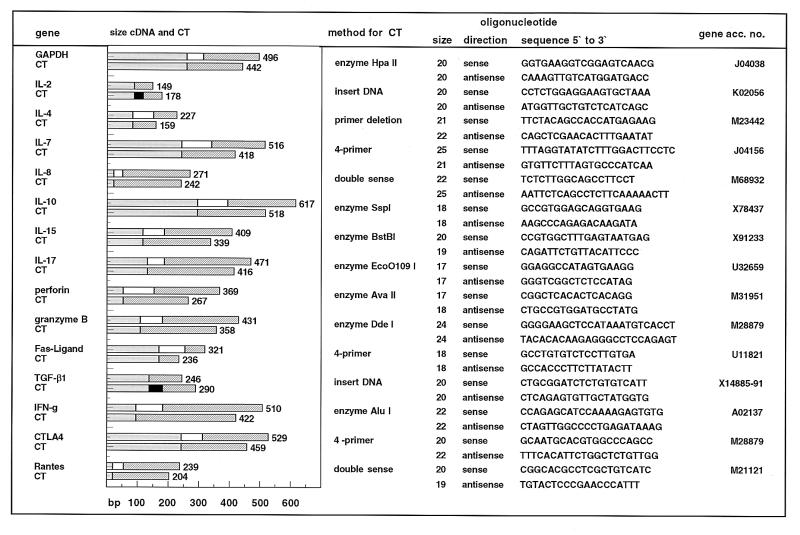
We have generated competitors from phytohemagglutinin-stimulated blasts or nephrectomy tissue by four different techniques (Fig. 1): (i) excision of a 50- to 100-bp fragment in the center of the target gene cDNA by using appropriate restriction enzymes (GAPDH, IFN-γ, IL-10, IL-15, IL-17, P, and GB); (ii) amplification of external parts of the cDNA by two separate PCRs and religation of these fragments (CTLA4, IL-7, FasL); (iii) insertion of a short DNA fragment into the target sequence (IL-2, IL-4, TGF-β1, or primer deletion (IL-4), kindly provided by M. Suthanthiran, Cornell Medical School, New York); and (iv) one-step generation of a shortened DNA sequence by use of a specifically designed double-sense primer. Competitors were cloned in a TA vector (Invitrogen, San Diego), transfected into DH5a cells (Promega), purified, and quantitated by UV spectrometry.
Figure 1.
Sequences of oligonucleotide primers and competitive templates (CTs) used for the quantitation of 15 genes evaluated. Deletions and insertions are indicated by black and white portions of the bars, respectively. The mutated competitors are coamplified with wild-type cDNA in PCR, and amplified wild-type and competitive template sequences are readily identified as distinct bands by agarose gel electrophoresis.
Amplification of the universally expressed GAPDH gene served to confirm successful RNA isolation and reverse transcription. The magnitude of target gene expression is calculated as pg of target gene cDNA per pg of GAPDH cDNA.
Statistical analysis was performed by the Beth Israel Hospital statistical bioanalysis core facility, using a Newman–Keuls test for normally distributed data or a Kruskal–Wallis test.
RESULTS
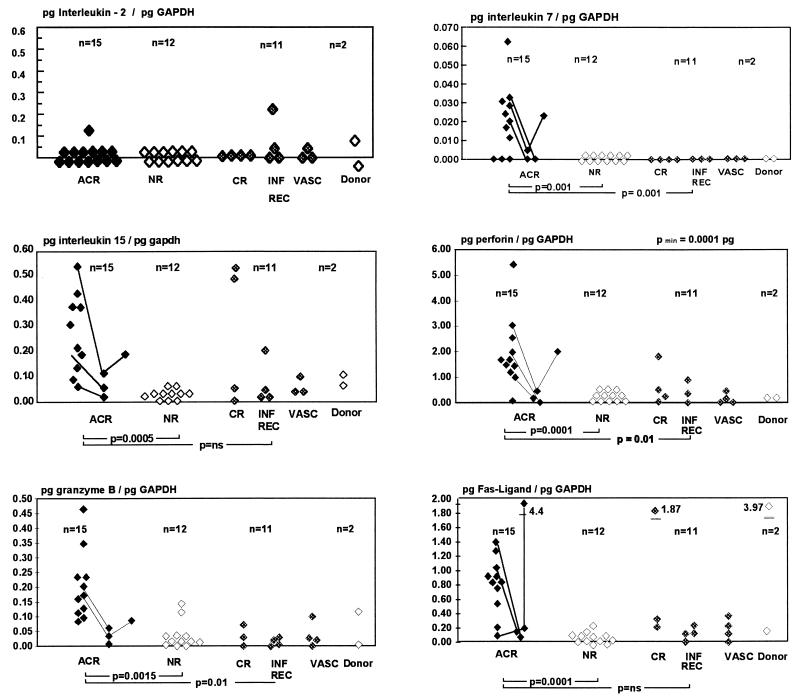
The small amount of tissue available for this study ((null)/1;10 to ½ of a biopsy core) proved to be sufficient for a thorough analysis of gene expression. The RNA yield ranged from 1 to 20 μg, depending on the size of the biopsy fragment, allowing 40–800 PCRs per sample. A quantitative analysis of gene expression for our purposes is a necessity, as low levels of transcripts are detectable in many biopsies, while heightened expression of select genes occurs only during rejection (Fig. 2, Table 1).
Figure 2.
Quantitative analysis of IL-2, IL-7, IL-15, P, GB, and FasL gene expression in 38 transplant core biopsies taken to aid in the differential diagnosis of graft dysfunction. Biopsies were also obtained from 2 donor kidneys prior to reperfusion. Lines indicate sequential biopsies taken during the course of rejection before and after treatment (ACR, acute cellular rejection; NR, nonrejecting kidneys with acute tubular necrosis or cyclosporine cytotoxicity; CR, chronic rejection; INF REC, infectious complications and recurrence of primary disease; and VASC, vascular complications).
Table 1.
Quantitative analysis of intragraft gene expression for 15 immune activation genes
| Gene | Rejection | Nonrejection | P* | Sensitivity, % | Specificity, % |
|---|---|---|---|---|---|
| IL-2 | 0.0 | 0.0 | NS | 8 | NA |
| IL-4 | 0.0 | 0.0 | NS | 0 | 0 |
| TGF-β1 | 112 ± 87 | 98 ± 78 | NS | 45 | 55 |
| CTLA-4 | 577 ± 396 | 228 ± 214 | <0.057 | 60 | 70 |
| RANTES | 284 ± 147 | 132 ± 104 | <0.064 | 91 | 71 |
| IFN-γ | 214 ± 194 | 151 ± 130 | 0.007 | 75 | 67 |
| IL-17 | 24 ± 12 | 0.0 | <0.001 | 83 | 75 |
| IL-7 | 38 ± 40 | 0.0 | <0.001 | 83 | 100 |
| IL-8 | 112 ± 82 | 67 ± | <0.0005 | 100 | 67 |
| IL-10 | 451 ± 340 | 24 ± 30 | <0.0005 | 83 | 89 |
| IL-15 | 236 ± 162 | 85 ± 37 | <0.0005 | 83 | 92 |
| GB | 174 ± 94 | 46 ± 51 | <0.0015 | 91 | 86 |
| P | 1705 ± 1021 | 338 ± 410 | <0.0001 | 83 | 92 |
| FasL | 779 ± 360 | 120 ± 101 | <0.0001 | 83 | 92 |
Values are given as mean ± SD pg of target gene cDNA per pg of GAPDH cDNA. The intensity of intragraft expression of individual CTL genes was compared with histologic (Banff) criteria for establishing the diagnosis of graft rejection through an analysis of 40 transplant biopsies and—in borderline cases—clinical response to antirejection treatment. NA, not applicable.
Statistical analysis was performed with a Newman–Keuls test for normally distributed data and a Kruskal–Wallis test for others. NS, not significant.
Heightened gene expression during acute rejection was detected for IL-7, IL-8, RANTES, IL-10, IL-15, IL-17, CTLA4, and all three CTL effector molecules—e.g., GB, P, and FasL (Fig. 2). GB and IL-10 expression (P < 0.0015 and P < 0.0005) proved to be significant and specific markers of acute, but not chronic, rejection, while IL-15 (P < 0.0015), FasL, and P (P < 0.0001 and P < 0.0001) transcription was augmented during acute allograft rejection and in some of the chronic rejection samples analyzed. The magnitude of expression of individual CTL-specific genes was not linked, and we found no evidence that the granula-dependent (GB, P) or the receptor-mediated (FasL) pathways are alternatively activated. IL-7 and IL-17 transcripts are solely, but not reliably, observed in rejecting samples, while an increase of IL-8 and RANTES mRNA can be found in both rejection and graft dysfunction related to other causes. The highest level of any target gene expression measured was 4.4 times higher than the amount of GAPDH gene expression in this sample (FasL in an acute rejection episode). IL-2 and IL-4 gene expression did not accompany rejection episodes.
The accuracy of this PCR-based molecular approach to verify rejection can be considerably enhanced by a simultaneous analysis of CTL gene expression (Table 2). If a discriminatory level for heightened gene expression is set to the mean ±95% confidence interval of values observed in nonrejecting kidneys (maximum 0.07 pg/pg of GAPDH for GB, 0.4 pg/pg of GAPDH for FasL, and 0.8 pg/pg of GAPDH for P), the combined analysis of all three CTL effector molecules identifies acute cellular rejection, including borderline cases with a sensitivity of 100% and a specificity of 100% in our series (P < 0.0001).
Table 2.
Combined analysis of CTL gene expression
| Gene | Rejection | Nonrejection | P* | Sensitivity, % | Specificity, % |
|---|---|---|---|---|---|
| P + GB one or both up-regulated | 11/12 | 5/28 | 0.00015* | 91 | 82 |
| FasL + GB one or both up-regulated | 12/12 | 4/28 | <0.0001* | 100 | 85 |
| FasL + GB + P any two up-regulated | 12/12 | 0/28 | <0.0001* | 100 | 100 |
Expression of an individual gene was deemed positive for values above the mean ± 95% confidence interval of nonrejecting kidneys (maximum 0.07 pg/pg of GAPDH for GB and 0.4 pg/pg of GAPDH for FasL and 0.8 pg/pg of GAPDH for P).
Statistical analysis was performed with a χ2 test.
The magnitude of gene expression indicative for those genes associated with rejection—i.e., GB, P, and FasL—apparently declines after initiation of effective antirejection therapy (OKT3 or steroid pulses) as exemplified in the few sequential biopsy specimens analyzed (Fig. 2).
Posttransplant surveillance biopsies showed similar levels of IL-7, IL-10, IL-17, and GB transcripts as compared with nonrejecting kidneys, while early (day 4 and 11) posttransplant specimens revealed that IL-15, CTLA4, P, and FasL mRNA levels were 2- 5-fold higher and showed a tendency to decline within the first week. In a limited sampling, early posttransplant gene expression was not predictive for the later development of rejection episodes.
DISCUSSION
Current diagnostic procedures in renal allograft rejection depend on detection of organ dysfunction and the subsequent histopathological examination. The typical clinical combination of azotemia and a mild cellular infiltration is often inconclusive, and the diagnosis of acute rejection has to be made retrospectively according to the response to antirejection therapy.
In agreement with our hypothesis that clinical allograft rejection is associated with expression of a specific subset of immune activation genes, we determined that allograft rejection is associated with intragraft expression of certain but not all T-cell-dependent activation genes.
Heightened gene expression of chemokines (IL-8, RANTES), non-T-cell-derived T-cell growth factors (IL-7, IL-15) and CTL-selective effector molecules was observed during rejection. The quantitative RT-PCR analysis of intragraft of IL-10 and IL-15 transcripts (macrophages) and the CTL-selective genes P, GB, and FasL provides a reliable and highly sensitive tool for the diagnosis of acute renal allograft rejection. RANTES and IL-8 transcripts proved to be sensitive but nonspecific indicators of rejection. IL-7 and IL-17 transcripts are seen only in rejection, but false negatives are commonplace. IL-2 and IL-4 gene expression is not detected in rejection samples, while expression of IFN-γ, TGF-β1, and CTLA4 genes was not selective for rejection.
A combined analysis of FasL, P, and GB heightens the diagnostic accuracy of this approach considerably as compared with an analysis of any individual gene transcript. Heightened gene expression of at least two of the three CTL genes is detected only in specimens from kidneys undergoing acute cellular rejection, while low expression of these genes was confined to biopsies with other causes of graft dysfunction.
Elevated IL-15, FasL, and P, but not IL-7, IL-10, or GB, transcripts were occasionally found in the few chronic rejection samples processed. This finding suggests a linkage between the causation of acute and chronic rejection.
The mere existence of a mononuclear leukocytic infiltrate, the hallmark for the histopathological diagnosis of rejection, may not necessarily be harmful for a transplant. Sequential biopsies obtained from well-functioning renal allografts at 3 and 6 months have frequently shown mononuclear leukocytic infiltrates (4, 5) without heightened expression for cytokines, P, or GB (19). Nonetheless some of these grafts have developed subsequent chronic rejection (5). In one experimental system, an effective cyclosporine regimen did not prevent graft infiltration, but such treatment lowered the frequency of CD8+ cells expressing P and G (21). In accordance with the notion that many graft-infiltrating T cells are not cytodestructive, in histological sections of rejecting human renal allografts only few T cells show P mRNA expression (18, 22). The small number of borderline cases examined in this study supports the idea that in case of a mild cellular infiltrate rejection can be identified by gene expression analysis.
Intragraft detection of CTL effector molecules may be a particularly sensitive diagnostic tool for acute rejection. GB and P have been shown to be excellent markers of acute allograft rejection at the mRNA and protein levels in animal models and clinical transplantation (12–18, 23, 24). This undoubtedly reflects differences in the functional programs of activated and resting lymphocytes. Activated, but not resting, CTLs and NK cells express FasL and GB. P can be detected in less than 30% of nonactivated CD8+ cells, while stimulation of peripheral blood mononuclear cells by anti-CD3 or IL-2 induces P expression in 60% of CD8+ cells and cytotoxic activity in CD8+ CD11b+ cells and γδ cells (25). FasL is detected primarily upon activated Th1 CD4+ cells, CD8+ CTLs (26), and NK cells (27) but is not expressed on macrophages or in renal tissue.
The functional role of heightened FasL expression in rejection remains to be determined. Although up-regulation of FasL transcripts has been noted in a murine heart transplant model, neither mutations affecting the Fas–FasL (20) genes nor targeted gene disruption for either GB or P has resulted in prolonged graft survival (28). Apoptosis has not been established as a major pathway of allograft injury, although DNA nick-end-labeling shows increased apoptosis in rejecting transplants (29–33). Transplantation of FasL-expressing tissue has resulted in prolonged graft survival, presumably through apoptotic elimination of graft-infiltrating Fas-bearing T and NK cells (34). Lynch et al. (35) have proposed a model of T-cell regulation where FasL provides an early costimulatory signal in CTL activation leading to IL-2-independent clonal expansion, while a Fas–FasL contact in later stages, when eliminated antigens no longer stimulate the T-cell receptor, provides a negative signal and leads to apoptosis of antigen-reactive T cells. In terms of this hypothesis, up-regulation of FasL transcripts in rejecting allografts indicates activated CTLs. While we are proposing that our study in no way implies that CTLs are essential or sufficient to mediate graft rejection, CTL gene expression provides an extraordinarily powerful diagnostic tool.
The effects of current posttransplant drug regimens on the dynamics of intragraft gene expression are unknown. Our data suggest that muted IL-7, IL-10, IL-15, and CTL gene expression may serve as an indicator for effective antirejection therapy (Fig. 2). Whether this effect occurs by gene regulation or cell elimination remains speculative. Corticosteroids and cyclosporine inhibit cytokine and FasL gene expression (36). Cyclosporine inhibits IL-2, IFN-γ, and FasL expression, presumably by blocking NF-κB activation (37) and up-regulating bcl-2 (38).
In clinical and experimental organ transplantation de novo T-cell gene expression precedes clinically detectable rejection. Through sequential evaluation of gene expression after transplantation we hope to identify and abrogate rejections in a nascent state. Our series of surveillance biopsies has not yet identified early markers of rejection.
It is interesting that IL-2 and IL-4 were not detected during rejection episodes. It will be of great interest to learn in an ongoing surveillance biopsy study whether (i) IL-2 gene expression precedes clinically evident rejection as noted in preclinical models (7) and (ii) IL-4 gene expression is detectable in long-term stable allografts. IL-4 gene expression frequently accompanies successful long-term engraftment in preclinical trials (39).
Recent modifications of our techniques, utilizing rapid RNA isolation methods, rigorous shortening of PCR cycler times, and multi-exon-spanning primers, allow us to quantify the expression of target genes in biopsies within a few hours. RT-PCR has been performed on RNA from fine needle aspirations (40, 41), peripheral blood, and urine sediments (42). In future studies it will be important to learn whether RT-PCR-detected gene expression events are accompanied by expression of the gene product through immunohistological analysis. All of these techniques offer a less invasive approach than core biopsies, but it remains to be determined whether the same reliability can be achieved. A potential limitation of any biopsy or aspiration-based technique relates to sampling errors, as early rejection is often patchy. PCR-based techniques do not obviate this problem.
While CTL gene analysis now provides a rapid and extraordinarily reliable tool to diagnose acute rejection even in cases with only mild cellular infiltrates, a future challenge will be to identify early warning markers and to utilize the sensitivity and specificity of RT-PCR to elucidate specific patterns of gene activation in vascular, chronic, and treatment-resistant rejections by refining the diagnostic criteria. It is our hope that these methods may, in the future, serve as a guide to the clinician in providing timely and specific therapy to prevent and treat rejection.
Acknowledgments
We thank Bernhard Ransell of the Beth Israel Hospital statistical bioanalysis core facility for providing skillfull and experienced help with statistical analysis of the large data base. This work was supported by grants from the Ronald McDonald Children’s Foundation Germany (J.S.), the Clinical Investigator Training Program, Beth Israel Hospital and Harvard/MIT Health Sciences and Technology, in collaboration with Pfizer Inc. (M.P.), the Medical Research Council of Canada (M.L.), and the Baxter Extramural Program and the National Institutes of Health (T.B.S.).
Footnotes
Abbreviations: NK, natural killer; IL, interleukin; IFN, interferon; CTL, cytotoxic T lymphocyte; GB, granzyme B; P, perforin; FasL, Fas ligand; RT, reverse transcription; TGF, transforming growth factor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Cecka, J. M. & Terasaki, P. I. (1993) Clin. Transplant. 1993, 1–18. [PubMed]
- 2.Tejani A, Stablein D, Alexander S, Fine R, Harmon W. Transplantation. 1995;59:500–504. [PubMed] [Google Scholar]
- 3.Solez K, Axelsen R A, Benediktsson H, Burdick J F, Cohen A H, Colvin R B, Croker R P, Droz D, Dunik M S, Halloran P F. Kidney Int. 1993;44:411–422. doi: 10.1038/ki.1993.259. [DOI] [PubMed] [Google Scholar]
- 4.Rush D N, Henry S F, Jeffery J R, Schroeder T J, Gough J. Transplantation. 1994;57:208–211. doi: 10.1097/00007890-199401001-00009. [DOI] [PubMed] [Google Scholar]
- 5.Rush D N, Jeffrey J R, Gough J. Transplantation. 1994;59:511–514. [PubMed] [Google Scholar]
- 6.Crabtree G R. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 7.O’Connell P J, Pacheco-Silva A, Nickerson P W, Muggia R A, Bastos M, Kelley V R, Strom T B. J Immunol. 1993;150:1093–1104. [PubMed] [Google Scholar]
- 8.Xu G P, Sharma V K, Li B, Bologa R, Li Y, Mouradian J, Wang J, Serur D, Rao V, Stenzel K H. Kidney Int. 1995;48:1504–1507. doi: 10.1038/ki.1995.440. [DOI] [PubMed] [Google Scholar]
- 9.Pavlakis M, Strehlau J, Lipman M, Shapiro M, Maslinski W, Strom T B. Transplantation. 1996;62:543–545. doi: 10.1097/00007890-199608270-00020. [DOI] [PubMed] [Google Scholar]
- 10.Strom T, Tilney N, Carpenter C, Busch G. New Engl J Med. 1975;292:1257–1263. doi: 10.1056/NEJM197506122922402. [DOI] [PubMed] [Google Scholar]
- 11.Bishop G A, Hall B M, Duggin G G, Horvath J S, Sheil A G, Tiller D J. Kidney Int. 1986;29:708–717. doi: 10.1038/ki.1986.56. [DOI] [PubMed] [Google Scholar]
- 12.Clement M V, Haddad P, Soulie A, Benvenuti C, Lichtenheld M G, Podak E R, Sigaux N, Sasportes M. Int Immunol. 1991;3:1175–1181. doi: 10.1093/intimm/3.11.1175. [DOI] [PubMed] [Google Scholar]
- 13.Legros-Maida S, Soulie A, Benvenuti C, Wargnier A, Vallee N, Berthouc C, Guillet J, Sasportes M, Sigaux N. Eur J Immunol. 1994;24:229–233. doi: 10.1002/eji.1830240136. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths G M, Namikawa R, Mueller C, Liu C C, Young J D, Billingham M, Weissman I. Eur J Immunol. 1991;21:687–693. doi: 10.1002/eji.1830210322. [DOI] [PubMed] [Google Scholar]
- 15.Clement M V, Legros-Maida S, Israel-Biet D, Carnot F, Soulie A, Reynaud P, Guillet J, Gandjbakch I, Sasportes M. Transplantation. 1994;57:322–326. doi: 10.1097/00007890-199402150-00002. [DOI] [PubMed] [Google Scholar]
- 16.Lipman M L, Stevens A C, Strom T B. J Immunol. 1994;152:5120–5127. [PubMed] [Google Scholar]
- 17.Kummer J A, Wever P C, Kamp A M, ten Berge I J, Hack C E, Weening J J. Kidney Int. 1995;47:70–77. doi: 10.1038/ki.1995.8. [DOI] [PubMed] [Google Scholar]
- 18.Matsuno T, Sakagami K, Saito S, Onoda T, Fujiwara T, Naomoto Y, Yagi T, Nakagawa H, Kusaka S, Orita K. Transplant Proc. 1992;24:1306–1307. [PubMed] [Google Scholar]
- 19.Lipman M, Jeffery J, Gough J, McKenna R, Grimm P, Rush D. J Am Soc Nephrol. 1995;6:1060. (abstr.) [Google Scholar]
- 20.Larsen C P, Alexander D Z, Hendrix R, Ritchie S C, Pearson T. Transplantation. 1995;60:221–224. doi: 10.1097/00007890-199508000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Mueller C, Shao Y, Andermatt H J, Hess M W, Shelby J. Transplantation. 1993;55:139–145. doi: 10.1097/00007890-199301000-00026. [DOI] [PubMed] [Google Scholar]
- 22.Grimm P C, McKenna R M, Gospodarek E M, Jeffery J R, Rush D N. Transplantation. 1995;59:579–584. [PubMed] [Google Scholar]
- 23.Sunder-Plassmann G, Wagner L, Hruby K, Balcke P, Worman C P. Kidney Int. 1990;37:1350–1356. doi: 10.1038/ki.1990.121. [DOI] [PubMed] [Google Scholar]
- 24.McDiarmid S V, Farmer D G, Kuniyoshi J S, Robert M, Khadavi A, Shaked A, Busuttil R W. Transplantation. 1995;59:762–766. [PubMed] [Google Scholar]
- 25.Nakata M, Kawasaki A, Azumi M, Tsuji K, Matsuda H, Shinkai Y, Yagita H, Okumura K. Int Immunol. 1992;4:1049–1054. doi: 10.1093/intimm/4.9.1049. [DOI] [PubMed] [Google Scholar]
- 26.Ramsdell F, Seaman M S, Miller R E, Picha K S, Kennedy M K, Lynch D H. Int Immunol. 1994;6:1545–1553. doi: 10.1093/intimm/6.10.1545. [DOI] [PubMed] [Google Scholar]
- 27.Arase H, Arase N, Saito T. J Exp Med. 1995;181:1235–1238. doi: 10.1084/jem.181.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz M, Schuurman H J, Joergensen J, Steiner C, Merloo T, et al. Eur J Immunol. 1995;25:474–480. doi: 10.1002/eji.1830250225. [DOI] [PubMed] [Google Scholar]
- 29.Nawaz S, Fennell R H. Histopathology. 1994;25:137–142. doi: 10.1111/j.1365-2559.1994.tb01569.x. [DOI] [PubMed] [Google Scholar]
- 30.Krams S M, Egawa H, Quinn M B, Martinez O M. Transplant Proc. 1995;27:466–467. [PubMed] [Google Scholar]
- 31.Tanaka K, Koga Y, Zhang X Y, Sasaki M, Wang Y, Kimura G, Nomoto K. J Immunol. 1993;151:748–758. [PubMed] [Google Scholar]
- 32.Matsuno T, Nakagawa K, Sasaki H, Ishine N, Inagaki M, Yagi T, Haisa M, Tanaka N, Sakagami K, Orita K. Transplant Proc. 1994;26:2170–2173. [PubMed] [Google Scholar]
- 33.Knoop M, McMahon R F, Jones C J, Hutchinson I V. Exp Pathol. 1991;41:219–224. doi: 10.1016/s0232-1513(11)80099-x. [DOI] [PubMed] [Google Scholar]
- 34.Bellgrau D, Gold D, Selawry H, Moore J, Franzusoff A, Duke R C. Nature (London) 1995;377:630–632. doi: 10.1038/377630a0. [DOI] [PubMed] [Google Scholar]
- 35.Lynch D H, Ramsdell F, Alderson M A. Immunol Today. 1995;16:569–574. doi: 10.1016/0167-5699(95)80079-4. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Mercep M, Ware C F, Ashwell J D. J Exp Med. 1995;181:1673–1682. doi: 10.1084/jem.181.5.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anel A, Buferne M, Boyer C, Schmitt-Verhulst A M, Golstein P. Eur J Immunol. 1994;24:2469–2476. doi: 10.1002/eji.1830241032. [DOI] [PubMed] [Google Scholar]
- 38.Labalette M, Queyrel V, Masy E, Noel C, Pruvot F R, Dessaint J P. Transplantation. 1995;59:1714–1723. doi: 10.1097/00007890-199506270-00013. [DOI] [PubMed] [Google Scholar]
- 39.Strom T B, Roy-Chaudhury P, Manfro R, Zheng X X, Nickerson P W, Wood K, Bushell A. Curr Opin Immunol. 1996;8:688–693. doi: 10.1016/s0952-7915(96)80087-2. [DOI] [PubMed] [Google Scholar]
- 40.Dallman M J, Larsen C P, Morris P J. J Exp Med. 1991;174:493–496. doi: 10.1084/jem.174.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nast C. Pediatr Nephrol NY. 1995;9:S56–S60. doi: 10.1007/BF00867686. [DOI] [PubMed] [Google Scholar]
- 42.Jeyarajah D R, Kadakia R A, O’Toole K, Newell K A, Josephson M A, Spargo B H, Woodle E S, Thistlewaite J R., Jr Transplant Proc. 1995;27:887–889. [PubMed] [Google Scholar]