Abstract
Hepatocyte growth factor/scatter factor (HGF/SF) is a mesenchymally derived, multifunctional paracrine regulator possessing mitogenic, motogenic, and morphogenetic activities in cultured epithelial cells containing its tyrosine kinase receptor, Met. c-met has been implicated in oncogenesis through correlation of expression with malignant phenotype in specific cell lines and tumors. Paradoxically, however, HGF/SF can also inhibit the growth of some tumor cells. To elucidate the oncogenic role of HGF/SF in vivo, transgenic mice were created such that HGF/SF was inappropriately targeted to a variety of tissues. HGF/SF transgenic mice developed a remarkably broad array of histologically distinct tumors of both mesenchymal and epithelial origin. Many neoplasms arose from tissues exhibiting abnormal development, including the mammary gland, skeletal muscle, and melanocytes, suggesting a functional link between mechanisms regulating morphogenesis and those promoting tumorigenesis. Most neoplasms, especially melanomas, demonstrated overexpression of both the HGF/SF transgene and endogenous c-met, and had enhanced Met kinase activity, strongly suggesting that autocrine signaling broadly promotes tumorigenesis. Thus, subversion of normal mesenchymal–epithelial paracrine regulation through the forced misdirection of HGF/SF expression induces aberrant morphogenesis and subsequent malignant transformation of cells of diverse origin.
Keywords: autocrine loop, breast cancer, malignant melanoma, Met, transgenic mice
Hepatocyte growth factor/scatter factor (HGF/SF) is characterized as a multifunctional cytokine based on its ability to stimulate proliferation, movement, and/or morphogenesis of a wide variety of cultured epithelial cells expressing the tyrosine kinase receptor encoded by the c-met protooncogene (1–6). Whereas HGF/SF is expressed in a multitude of mesenchymally derived cells, Met expression has been detected in the epithelium of almost all tissues, indicating that under normal physiological conditions HGF/SF functions almost exclusively as a paracrine regulator (7, 8). During development, HGF/SF is expressed in distinct mesenchymal embryonic tissues lying in close proximity to Met-expressing epithelium, including the olfactory mucosa of the nasal cavities, somitic myogenic precursors, and the embryonic spinal cord, suggesting that the HGF/SF–Met signal transduction pathway helps mediate mesenchymal–epithelial interactions in vivo (9, 10). In addition, it has been shown in vitro that paracrine HGF/SF stimulation is required for appropriate mammary gland development (11, 12). Recent genetic studies have directly demonstrated a requirement for the HGF/SF-Met signaling pathway in normal development. Mouse embryos carrying null mutations in both HGF/SF alleles demonstrated impaired liver and placental development and died in midgestation (13, 14). Furthermore, c-met null mutant mouse embryos had skeletal muscle defects of the limbs and diaphragm, indicating that Met plays an important role in skeletal muscle development (15).
Met has also been implicated in oncogenesis. c-met was first identified as a protooncogene through its ability to transform NIH 3T3 cells as a rearranged gene fusion in which c-met coding sequences were juxtaposed with the translocated promoter region (16, 17). c-met has also been shown to be inappropriately expressed in diverse human and mouse tumors including melanomas, rhabdomyosarcomas, hepatomas, and carcinomas of the breast (18–26). The c-met gene is amplified in some carcinomas of the gastrointestinal tract (22, 27). Moreover, coexpression of HGF/SF and Met has been identified in a variety of transformed cultured cells and in some tumors (23, 28–30). In some cases HGF/SF–Met overexpression has been correlated with tumor progression and/or metastasis (20, 24–26, 28). Together, these studies have suggested that the creation of HGF/SF–Met autocrine loops can be intimately associated with neoplastic transformation. Paradoxically, however, HGF/SF has also been reported to inhibit the growth of certain carcinoma cell lines (31, 32).
To elucidate the in vivo role of HGF/SF in tumorigenesis, we have generated transgenic mice in which robust expression of a mouse HGF/SF cDNA was broadly targeted to a wide variety of tissues using the mouse metallothionein (MT) gene promoter and 5′ and 3′ genomic flanking sequences. Previously, we reported that these transgenic mice exhibit ectopic skeletal muscle and melanocytes in the central nervous system, suggesting that HGF/SF possesses scatter activity in vivo and can function as a true morphogenetic factor by regulating migration and/or differentiation of select populations of myoblasts and neural crest cells during embryogenesis (33). Here we show that the establishment of HGF/SF–Met autocrine signaling loops induces diverse tumorigenesis in a wide variety of tissue types, many of which demonstrate associated developmental abnormalities.
MATERIALS AND METHODS
Transgenic Mice.
MT–HGF/SF transgenic mice were generated on an albino FVB/N background as described (33). An MT-1 promoter was chosen to drive expression of a mouse HGF/SF cDNA. MT 5′ and 3′ flanks were included that contain locus control regions conferring copy number-dependent and position-independent transgene expression (33, 34). Mouse work was performed in accordance with National Institutes of Health guidelines.
Analysis of Tissues and Tumors.
Tissues were routinely fixed in 10% buffered formalin, paraffin-embedded, sectioned at five microns, and stained with hematoxylin and eosin. For electron microscopy, tissues were fixed in freshly prepared glutaraldehyde. mHGF/SF was localized using a rabbit anti-human polyclonal antibody as suggested by the manufacturer (R&D Systems). In some cases, tumor classification was confirmed with the aid of immunohistochemistry and/or electron microscopy. Rhabdomyosarcomas were positively identified by α-actin immunostaining (Dako). All amelanotic melanomas were identified by their histomorphological appearance, which resembled melanomas found in rats (35), and by positive immunostaining with S-100 (Sigma). In addition, candidate melanomas were confirmed by positive immunostaining with HMB45, a monoclonal antibody reactive with human immature melanosomes and melanoma tumor cells (Dako), electron microscopy, and/or tyrosinase gene expression by Northern blot hybridization (not shown). Statistical analysis of tumor incidence was by the Fisher’s exact test.
Analysis of RNA and DNA.
HGF/SF transcripts were detected by Northern blot hybridization using a PCR-generated probe (33). Total tissue RNA was isolated as described (36), and 20 μg were loaded per lane on an agarose gel to resolve RNA transcripts. The HGF/SF cDNA probe was synthesized by PCR as described (33). A mouse c-met cDNA hybridization probe was prepared by PCR from a 1.5-kb EcoRI–EcoRI fragment subcloned from the c-met cDNA clone C1 (37). On occasion, the MT transgene promoter was maximally induced by exposure to zinc, either in drinking water containing 25 mM zinc sulfate or by injection of 10 mg/kg body weight zinc chloride 5 hr before death. To control for RNA loading and transfer variation, filters were routinely rehybridized with a glyceraldehyde-3-phosphate dehydrogenase probe.
Amplification of c-met was assessed by Southern blot hybridization. Genomic DNA samples were digested with EcoRI, transferred to nitrocellulose, and probed with the 1.5-kb c-met cDNA as described above.
Analysis of Met and Met Activation.
Quantification of Met and Met tyrosine phosphorylation was basically as described (38). Five micrograms of whole tissue (wet weight equivalents) were solubilized in RIPA buffer (50 mM Tris, pH 7.4/50 mM NaCl/1.0% Triton X-100/5 mM EDTA/10 mM sodium pyrophosphate/50 mM sodium fluoride/1 mM sodium orthovanadate/1 mM phenylmethylsulfonyl fluoride/10 μg/ml leupeptin/10 μg/ml pepstatin/1 μg/ml aprotinin). Equivalent amounts of cleared lysate were incubated with phosphotyrosine monoclonal antibody (Upstate Biotechnology, Lake Placid, NY), control rabbit antibody, or anti-Met antibody (Santa Cruz Biotechnology) for 2 hr. Following addition of protein A-Sepharose CL4B, and washing in RIPA buffer, samples were fractionated on reducing SDS/8% polyacrylamide gels. After electrophoretic transfer to Immobilon P membranes, filters were blocked and then incubated with anti-Met antibody overnight. Met was visualized by incubation with anti-rabbit antibody conjugated to horseradish peroxidase and by using enhanced chemiluminescence (Amersham).
RESULTS
Appearance of Diverse Histological Tumor Types in HGF/SF Transgenic Mice.
Mice have been generated in which an HGF/SF transgene was overexpressed relative to the endogenous gene by a factor of between 3- and 50-fold, as described (33). Six independently derived lines of HGF/SF transgenic mice developed multiple tumor types between 2.5 and 20 months of age whose diversity was striking. Neoplasms arose from cells of both epithelial and mesenchymal origin, and from a wide range of tissues, including the skin, mammary and olfactory glands, liver, and muscle (see Fig. 1, Tables 1 and 2 for details).
Figure 1.
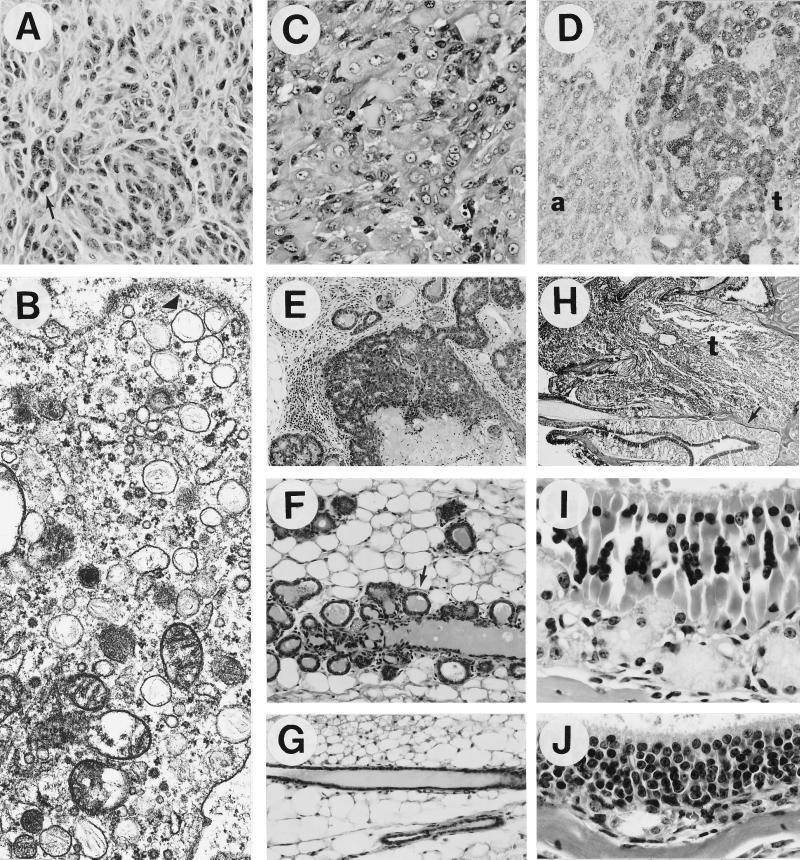
Aberrant morphogenesis and oncogenesis in HGF/SF transgenic mice. (A) Primary malignant amelanotic melanoma that metastasized widely. Note whorling pattern and multiple mitotic figures (arrow). (B) Electron micrograph of a melanoma cell with numerous melanosomes (arrowhead indicates a cluster). (C) Rhabdomyosarcoma that arose in the skin; note multiple mitotic figures (arrow). (D) Strong HGF/SF staining in a liver tumor (t) relative to adjacent tissue (a). (E) Characteristic mammary adenosquamous carcinoma composed of both mammary and hair follicle elements. (F) Precocious development in transgenic virgin females of alveolar structures (arrow), which expressed casein and whey acidic protein (G. Smith and G.M., unpublished data). (G) Normal ducts in nontransgenic virgin females. (H) Olfactory adenocarcinoma (t), which obliterated one side of the nasal olfactory mucosa; unaffected but disorganized olfactory mucosa is indicated by the arrow. (I) Higher magnification of the disorganized transgenic olfactory mucosa with degenerated epithelium, marked hypertrophy and hyperplasia of glands, and depleted nerves. Malformation of the nerves was confirmed by immunohistochemical staining using an antibody to S-100 (data not shown). (J) Nontransgenic olfactory mucosa. (A and C, ×400; B, ×39,500; D, F, and G, ×200; E, ×100; H, ×25; I and J, ×630.)
Table 1.
Incidence of selected tumors in HGF/SF transgenic mice
| Mouse genotype | Total mice | Mean age, months | Mammary tumors | Amelanotic melanomas | Rhabdo- myosarcomas | |
|---|---|---|---|---|---|---|
| Female | TG | 27 | 15.2 ± 4.1 | 11 (41%)* | 1 (4%) | 0 (0%) |
| WT | 29 | 13.0 ± 5.6 | 0 (0%) | 0 (0%) | 0 (0%) | |
| Male | TG | 42 | 15.3 ± 4.1 | 0 (0%) | 9 (21%)† | 3 (7%)‡ |
| WT | 25 | 17.3 ± 3.8 | 0 (0%) | 0 (0%) | 0 (0%) |
Tumor incidence is based on analysis of transgenic (TG) and wild-type (WT) mice >6 months of age. In TG mice <6 months of age, 3% had melanomas and 5% rhabdomyosarcomas. The presence or absence of tumors was based on gross inspection at necropsy. Tumors were classified by a variety of methods (see Materials and Methods). Liver tumor incidence also was significantly enhanced in aged TG mice; those data are presented elsewhere (39). Mean age is shown in months (±SD).
Mammary tumors arose in both virgin and parous females, and include adenocarcinomas and adenosquamous carcinomas with hair follicle and mammary gland elements (see text). P < 0.0005.
One of nine melanomas was metastatic to multiple organs. P < 0.05.
One of these three rhabdomyosarcomas was metastatic to the pancreas.
Table 2.
Characterization of representative tumors from HGF/SF transgenic mice
| Tumor no. | Age, months | TG line | Sex | Histological classification | Tissue of origin |
|---|---|---|---|---|---|
| 1 | 10 | 37 | M | Amelanotic melanoma* | Skin |
| 4 | 12 | 22 | F | Adenocarcinoma | Mammary |
| 33 | 13 | 22 | F | Hair follicle tumor | Skin |
| 38 | 16 | 22 | M | Rhabdomyosarcoma | Skin |
| 39 | 18 | 22 | F | Adenosquamous carcinoma | Mammary |
| 40 | 12 | 22 | F | Adenosquamous carcinoma | Mammary |
| 41 | 17 | 37 | M | Amelanotic melanoma | Exorbital |
| 47 | 20 | 31 | M | Hemangiosarcoma | Liver |
| 62 | 5 | 19 | F | Fibrosarcoma | Skin |
| 70 | 4 | 37 | M | Rhabdomyosarcoma | Skin |
| 79 | 15 | 22 | F | Adenocarcinoma | Mammary |
| 82 | 14 | 18 | M | Fibrosarcoma | Skin |
| 91 | 14 | 22 | M | Adenocarcinoma | Salivary |
| 93 | 17 | 37 | M | Hepatocellular adenoma | Liver |
| 95 | 11 | 37 | M | Amelanotic melanoma | Skin |
| 99 | 14 | 27 | F | Adenocarcinoma | Mammary |
Description of tumors (identified by number) arising in HGF/SF transgenic (TG) mouse lines subjected to molecular and biochemical analyses shown in Fig. 2. In addition to the tumor types listed above, the following were detected in one or more HGF/SF transgenic mice: adrenal cortical adenoma, basal cell tumor, clitoral gland adenoma, harderian gland adenoma, hemangioma, hepatocellular carcinoma, histiocytic sarcoma, metastatic lymphoblastic lymphoma, olfactory gland adenocarcinoma, squamous cell papilloma, and thyroid follicular cell adenoma. M, male; F, female.
Malignant with metastases.
The most prevalent tumors arose in the female mammary gland (Table 1). Forty-one percent of transgenic females over 6 months of age, virgin or parous, developed malignant mammary tumors that were of two types: adenocarcinoma and adenosquamous carcinoma. The latter had multifocal areas of squamous metaplasia that closely resembled hair follicle tumors (pilomatrixoma, Fig. 1E). Tumor incidence in the mammary gland of the parent FVB/N line was very low, as reported previously (40).
The skin, which potently expresses the HGF/SF transgene (33) and contains many Met-expressing cell types (41), was a particularly rich source of tumors. These included melanomas, rhabdomyosarcomas, fibrosarcomas, squamous papillomas, basal cell and hair follicle tumors. Twenty-one percent of transgenic males over 6 months of age developed amelanotic melanomas in the skin and subcutaneum (Table 1). Amelanotic melanomas were diagnosed based on a combination of histomorphology, immunohistochemistry, electron microscopy, and/or tyrosinase gene expression. Morphologically, melanomas were either of epithelioid cell type similar to pigmented melanomas in rats and other species or spindle cell type resembling schwannomas with the Antonini type A pattern as reported in albino Fischer 344 rats (35), or of a mixture of both. One subcutaneous melanoma with such a mixture (Fig. 1 A and B) was particularly aggressive and highly metastatic to multiple distant organs such as liver, pancreas, spleen, epididymis, and lymph nodes, and was readily transplantable to syngeneic FVB/N mice (data not shown). Another highly malignant tumor was a rhabdomyosarcoma that arose in the peritoneum and was metastatic to the pancreas.
Coexpression of HGF/SF and Met in Tumors of Mesenchymal and Epithelial Origin.
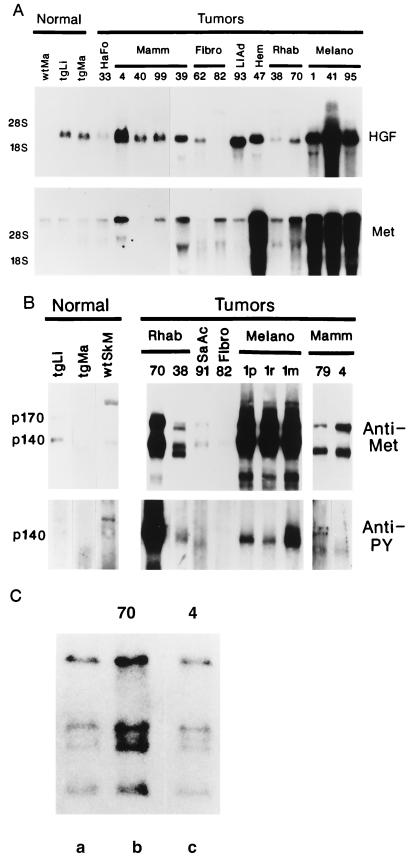
Representative transgenic tumors, described in Table 2, were subjected to molecular and/or biochemical characterization. Fig. 2A shows that transgenic HGF/SF RNA transcripts were abundant in most tumors analyzed. Occasionally, more HGF/SF protein could be detected in tumors relative to surrounding tissue; an HGF/SF-positive liver tumor is shown in Fig. 1D. Endogenous c-met transcripts were also present in most tumors examined. In more than one-half of these tumors c-met RNA levels were elevated, especially in the melanomas and the hemangiosarcoma where transcript levels were dramatically enhanced (Fig. 2A).
Figure 2.
Molecular and biochemical analyses of HGF/SF and c-met in transgenic neoplasms. (A) Northern blot detection of transgenic HGF/SF (Upper) and c-met (Lower) RNA transcripts in tumors and normal tissue. Migration of 28S and 18S rRNAs are shown at left. (B) Quantification of Met protein by anti-Met immunoprecipitation followed by anti-Met immunoblotting (Upper) and Met kinase activity by anti-phosphotyrosine (PY) immunoprecipitation followed by anti-Met immunoblotting (Lower). Location of p140 and p170 forms of Met are shown at left. Note greatly enhanced kinase activity associated with rhabdomyosarcoma 70. Normal tissues: wtMa, wild-type mammary; wtSkM, wild-type skeletal muscle; tgLi, transgenic liver; tgMa, transgenic mammary. Neoplasms: HaFo, hair follicle tumor; Hem, hemangiosarcoma; Mamm, mammary tumors; Melano, melanoma; Fibro, fibrosarcoma; LiAd, liver adenoma; Rhab, rhabdomyosarcoma; SaAc, salivary adenocarcinoma. The 1p, 1r, and 1m labels designate samples from primary, recurrent (following resection), and metastatic melanoma number 1. (C) Amplification of c-met DNA in overexpressing rhabdomyosarcoma 70 (lane b), but not in overexpressing mammary adenocarcinoma 4 (lane c). (Lane a) Unamplified c-met sample. Numbers throughout identify specific tumors (see Table 2).
Met protein levels were analyzed by immunoprecipitation and immunoblotting with anti-Met antibody. The p140 and presumably nonproteolytically processed p170 forms of Met protein were present in most tumors at levels that were consistent with c-met RNA expression (Fig. 2B). Met was often highly abundant, suggesting that Met overexpression is important in tumor formation and/or progression in this transgenic model system. Fig. 2C shows that in at least one rhabdomyosarcoma (shown in Fig. 1C) a 3-fold amplification in c-met contributed to strong overexpression.
Autocrine Met Activation as the Mechanistic Basis for Tumor Formation in Diverse Transgenic Tissues.
To investigate the mechanism by which HGF/SF acted as an oncogenic agent, we quantified Met tyrosine phosphorylation in various tumors. Met association with tyrosine phosphorylation was analyzed by immunoprecipitation with an anti-phosphotyrosine antibody followed by immunoblotting with anti-Met antibody. Fig. 2B shows that in fact p140 Met was associated with tyrosine phosphorylated complexes in most tumors. Kinase activity roughly correlated with Met levels; the greatest phosphorylation was associated with the metastatic melanoma and the rhabdomyosarcoma harboring amplified c-met. Kinase activity in this rhabdomyosarcoma (tumor number 70) appeared to be disproportionally high relative to expression of HGF/SF and Met (Fig. 2B). Tyrosine phosphorylated p170 Met was not detectable in any tissue tested, consistent with previous observations that this nonprocessed precursor does not bind the HGF/SF ligand (42). Our results show that in this in vivo model, diverse tumorigenesis was driven by an HGF/SF–Met autocrine loop.
Transgenic Tissues Predisposed to Tumorigenesis Exhibited Abnormal Development.
Significantly, tumors frequently arose from transgenic tissues exhibiting morphogenetic abnormalities, including muscle (rhabdomyosarcoma) and neural crest (melanoma), which had already been shown to develop at ectopic sites (33). In addition, the transgenic mammary gland, which expressed high levels of HGF/SF RNA (33) and was prone to the formation of adenocarcinomas and adenosquamous carcinomas, demonstrated incomplete penetration of ductal epithelium into the mesenchymal fat pad and precocious formation of alveolar structures in virgin females (Fig. 1 F and G). Transplantation of fragments of HGF/SF transgenic mammary epithelium into depopulated fat pads of FVB/N nontransgenic mice resulted in the focal appearance of the same atypical phenotype (G. Smith and G.M., unpublished results), suggesting that the observed developmental abnormalities were at least in part intrinsic to the transgenic mammary gland.
In all HGF/SF transgenic mice the structure of the olfactory mucosa, which also demonstrated the potential to form tumors, was found to be highly disorganized and to exhibit marked degeneration; olfactory epithelial cells and nerve bundles were often depleted, while olfactory glands became more prominent (Fig. 1 I and J). Although more severe in adult mice, similar changes were found in juvenile mice and in neonates, suggesting that these abnormalities were developmental in nature.
DISCUSSION
HGF/SF–Met autocrine signaling has been implicated in oncogenesis since a number of tumors coexpress HGF/SF and Met, and because Met autocrine activation can transform cells in culture (19, 23, 24, 28, 29, 43–45). To strictly test this correlation and to elaborate the oncogenic potential of HGF/SF in vivo, we decided to employ transgenic mice as a model system. In this study, we show that targeting expression of an HGF/SF transgene to most tissue types induces the development of a remarkably wide variety of histologically distinct neoplasms of both mesenchymal and epithelial origin. Moreover, most tumors coexpressed both HGF/SF and Met and had Met kinase activity, indicating an association between the creation of HGF/SF–Met autocrine loops and oncogenesis. This association was particularly compelling in the case of the melanomas, which demonstrated very high expression of both the HGF/SF transgene and c-met, and occasionally a metastatic phenotype.
These results may have relevance to mechanisms associated with the development of human cancer. Both human melanomas and rhabdomyosarcomas have been reported to overexpress Met, which was correlated with tumor progression and invasiveness (19, 20, 25). And although loss of heterozygosity at chromosome 7q (to which genes encoding both HGF/SF and Met map) has been correlated with human breast cancer and with shorter patient metastasis-free and survival times (46), coexpression of HGF/SF and Met has been detected in breast carcinomas (23), and high levels of immunoreactive HGF/SF in tumor extracts has been proposed to be an important prognostic factor in breast cancer (21). Our transgenic mice developed melanomas, rhabdomyosarcomas, and mammary tumors, many characterized by the creation of HGF/SF–Met autocrine loops, strongly supporting the notion that deregulated Met signal transduction can play an important role in human cancer. This conclusion is further supported by a very recent report showing that expression of the tpr-met oncogene induces mammary carcinogenesis in transgenic mice (47).
Met was highly abundant and active in many transgenic tumors, suggesting that HGF/SF transgenic cells overexpressing Met gain a selective growth advantage during tumor formation and/or progression. We have shown that amplification of the endogenous c-met gene can contribute to receptor overexpression in some neoplasms, as has been demonstrated in human gastrointestinal tumors (22, 27). Alternatively, another possibility is that elevated levels of HGF/SF itself induce c-met overexpression, as has been described (48). Finally, the disproportionally high levels of phosphotyrosine associated with one rhabdomyosarcoma (tumor number 70, Fig. 2B) raise the possibility that mutations in c-met elevate receptor kinase activity. In contrast to the up-regulation of endogenous c-met expression observed in HGF/SF transgenic tumors, liver and mammary tumors that developed in transforming growth factor α transgenic mice demonstrated unchanged or decreased epidermal growth factor receptor expression (49, 50). This underscores the possibility that differential mechanisms operate in creating specific ligand–receptor autocrine loops associated with tumorigenesis.
Significantly, tumors often arose from transgenic tissues that exhibited developmental abnormalities, raising the possibility that the two phenotypes may be functionally related. Transgenic mice had ectopic skeletal muscle in the central nervous system and inappropriate melanocyte localization to aberrant sites, suggesting that HGF/SF helps regulate the migration and/or differentiation of premyogenic and premelanocytic cells during embryogenesis (33). Autocrine expression of HGF/SF and Met is normally restricted to gastrulation and early organogenesis in the mouse, occurring in some incompletely differentiated migratory cell types including condensing somites and neural crest (51). Forced autocrine Met stimulation may therefore favor the maintenance of the undifferentiated state in skeletal muscle and melanocyte precursors, creating potential targets for malignant transformation and leading to the development of rhabdomyosarcomas and melanomas. Alternatively, cells that either migrate to or develop at ectopic sites could be removed from their normal milieu and any paracrine regulatory mechanisms associated therein, encouraging abnormal growth and subsequent tumorigenesis.
As another example, the transgenic mammary gland, which expressed high levels of HGF/SF RNA (33) and was prone to tumor formation, exhibited incomplete penetration of ductal epithelium into the mesenchymal fat pad and precocious formation of alveolar structures containing cells with mitotic figures in virgin females. Since endogenous HGF/SF and Met are known to be expressed in the mammary fat pad and epithelial cells, respectively (52), the perturbation of normal morphogenesis observed in transgenic mammary glands strongly suggests that HGF/SF helps mediate critical mesenchymal–epithelial paracrine communications during normal glandular development which, when subverted by inappropriate HGF/SF expression and autocrine Met activation, contribute to uncontrolled cellular growth and oncogenesis. A similar conclusion can be reached concerning the olfactory mucosa, in which Met-containing epithelium also normally develops in close proximity to HGF/SF-secreting mesenchyme (9). The olfactory mucosa was highly disorganized in transgenic neonatal and adult mice, and characterized by epithelial degeneration, nervous depletion, glandular hyperplasia and hypertrophy, and adenocarcinoma formation.
It has long been appreciated that neoplastic and undifferentiated multipotent cells share important properties. Here we show that HGF/SF overexpression induces the formation of a broad spectrum of sarcomas and carcinomas demonstrating autocrine Met activation, many arising from tissues exhibiting striking developmental abnormalities. Our results underscore the importance of paracrine signaling in regulating normal cellular processes such as growth, differentiation, and migration, and demonstrate the pathological consequences of perturbing that signaling. Since analyses of both HGF/SF and c-met null mice have been hampered by early embryonic lethalities (13–15), our HGF/SF transgenic mice represent a valuable tool to elucidate the underlying mechanisms of mesenchymal–epithelial interaction and their relationship to morphogenesis and oncogenesis.
Acknowledgments
We thank Dr. Jennie Owens for electron microscopy; Drs. Jerrold Ward, Jackie Pierce, Hiromi Sakata, Jeffrey Rubin, Ira Pastan, and Gilbert Smith for useful discussions; Dr. Richard Palmiter for the MT locus control region clones; and Ricardo Dreyfuss and Steve Neal for photography.
Footnotes
Abbreviations: HGF/SF, hepatocyte growth factor/scatter factor; MT, metallothionein.
References
- 1.Gherardi E, Stoker M. Cancer Cells. 1991;3:227–232. [PubMed] [Google Scholar]
- 2.Birchmeier C, Sonnenberg E, Weidner K M, Walter B. BioEssays. 1993;15:185–190. doi: 10.1002/bies.950150307. [DOI] [PubMed] [Google Scholar]
- 3.Rosen E M, Nigam S K, Goldberg I D. J Cell Biol. 1994;127:1783–1787. doi: 10.1083/jcb.127.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarnegar R, Michalopoulos G K. J Cell Biol. 1995;129:1177–1180. doi: 10.1083/jcb.129.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkmann V, Foroutan H, Sachs M, Weidner K M, Birchmeier W. J Cell Biol. 1995;131:1573–1586. doi: 10.1083/jcb.131.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boros P, Miller C M. Lancet. 1995;345:293–295. doi: 10.1016/s0140-6736(95)90279-1. [DOI] [PubMed] [Google Scholar]
- 7.Iyer A, Kmiecik T E, Park M, Daar I, Blair D, Dunn K J, Sutrave P, Ihle J N, Bodescot M, Vande Woude G F. Cell Growth Differ. 1990;1:87–95. [PubMed] [Google Scholar]
- 8.Di Renzo M F, Narsimhan R P, Olivero M, Bretti S, Giordano S, Medico E, Gaglia P, Zara P, Comoglio P M. Oncogene. 1991;6:1997–2003. [PubMed] [Google Scholar]
- 9.Sonnenberg E, Meyer D, Weidner K M, Birchmeier C. J Cell Biol. 1993;123:223–235. doi: 10.1083/jcb.123.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsarfaty I, Rong S, Resau J H, Rulong S, da Silva P P, Vande Woude G F. Science. 1994;263:98–101. doi: 10.1126/science.7505952. [DOI] [PubMed] [Google Scholar]
- 11.Soriano J V, Pepper M S, Nakamura T, Orci L, Montesano R. J Cell Sci. 1995;108:413–430. doi: 10.1242/jcs.108.2.413. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Spitzer E, Meyer D, Sachs M, Niemann C, Hartmann G, Weidner K M, Birchmeier C, Birchmeier W. J Cell Biol. 1995;131:215–226. doi: 10.1083/jcb.131.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N. Nature (London) 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Nature (London) 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 15.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Nature (London) 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 16.Park M, Dean M, Cooper C S, Schmidt M, O’Brien S J, Blair D G, Vande Woude G F. Cell. 1986;45:895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- 17.Dean M, Park M, Vande Woude G F. Mol Cell Biol. 1987;7:921–924. doi: 10.1128/mcb.7.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prat M, Narsimhan R P, Crepaldi T, Nicotra M R, Natali P G, Comoglio P M. Int J Cancer. 1991;49:323–328. doi: 10.1002/ijc.2910490302. [DOI] [PubMed] [Google Scholar]
- 19.Rong S, Jerrers M, Resau J H, Tsarfaty I, Oskarsson M, Vande Woude G F. Cancer Res. 1993;53:5355–5360. [PubMed] [Google Scholar]
- 20.Natali P G, Nicotra M R, Di Renzo M R, Prat M, Bigotti A, Cavaliere R, Comoglio P M. Br J Cancer. 1993;68:746–750. doi: 10.1038/bjc.1993.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita J, Ogawa M, Yamashita S, Nomura K, Kuramoto M, Saishoji T, Shin S. Cancer Res. 1994;54:1630–1633. [PubMed] [Google Scholar]
- 22.Di Renzo M F, Olivero M, Giacomini A, Porte H, Chastre E, Mirossay L, Nordlinger B, Bretti S, Bottardi S, Giordano S, Plebani M, Gespach C, Comoglio P M. Clin Cancer Res. 1995;1:147–154. [PubMed] [Google Scholar]
- 23.Tuck A B, Park M, Sterns E E, Boag A, Elliott B E. Am J Pathol. 1996;148:225–232. [PMC free article] [PubMed] [Google Scholar]
- 24.Rong S, Donehower L A, Hansen M F, Strong L, Tainsky M, Jeffers M, Resau J H, Hudson E, Tsarfaty I, Vande Woude G F. Cancer Res. 1995;55:1963–1970. [PubMed] [Google Scholar]
- 25.Ferracini R, Olivero M, Di Renzo M F, Martano M, De Giovanni C, Nanni P, Basso G, Scotlandi K, Lollini P-L, Comoglio P M. Oncogene. 1996;11:1697–1705. [PubMed] [Google Scholar]
- 26.Di Renzo M F, Olivero M, Ferro S, Prat M, Bongarzone I, Pilotti S, Belfiore A, Constantio A, Vigneri R, Pierotti M A, Comoglio P M. Oncogene. 1992;7:2549–2553. [PubMed] [Google Scholar]
- 27.Kuniyasu H, Yasui W, Kitadai Y, Yokozaki H, Ito H, Tahara E. Biochem Biophys Res Commun. 1992;189:227–232. doi: 10.1016/0006-291x(92)91548-5. [DOI] [PubMed] [Google Scholar]
- 28.Bellusci S, Moens G, Gaudino G, Comoglio P, Nakamura T, Thiery J-P, Jouanneau J. Oncogene. 1994;9:1091–1099. [PubMed] [Google Scholar]
- 29.Rong S, Segal S, Anver M, Resau J H, Vande Woude G F. Proc Natl Acad Sci USA. 1994;91:4731–4735. doi: 10.1073/pnas.91.11.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahimi N, Tremblay E, McAdam L, Park M, Schwall R, Elliott B. Cell Growth Differ. 1996;7:263–270. [PubMed] [Google Scholar]
- 31.Tajima H, Matsumoto K, Nakamura T. FEBS Lett. 1991;291:229–232. doi: 10.1016/0014-5793(91)81291-f. [DOI] [PubMed] [Google Scholar]
- 32.Shiota G, Rhoads D B, Wang T C, Nakamura T, Schmidt E V. Proc Natl Acad Sci USA. 1992;89:373–377. doi: 10.1073/pnas.89.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takayama H, LaRochelle W J, Anver M, Bockman D, Merlino G. Proc Natl Acad Sci USA. 1996;93:5866–5871. doi: 10.1073/pnas.93.12.5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmiter R D, Sandgren E P, Koeller D M, Brinster R L. Mol Cell Biol. 1993;13:5266–5275. doi: 10.1128/mcb.13.9.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshitomi K, Elwell M R, Boorman G A. Toxicol Pathol. 1995;23:16–25. doi: 10.1177/019262339502300103. [DOI] [PubMed] [Google Scholar]
- 36.Jhappan C, Stahle C, Harkins R N, Fausto N, Smith G H, Merlino G T. Cell. 1990;61:1137–1146. doi: 10.1016/0092-8674(90)90076-q. [DOI] [PubMed] [Google Scholar]
- 37.Chan A M-L, King H W S, Deakin E A, Tempest P R, Hilkens J, Kroezen V, Edwards D R, Wills A J, Brooks P, Cooper C S. Oncogene. 1988;2:593–599. [PubMed] [Google Scholar]
- 38.LaRochelle W J, Jensen R A, Heidaran M A, May-Siroff M, Wang L M, Aaronson S A, Pierce J H. Cell Growth Differ. 1993;4:547–553. [PubMed] [Google Scholar]
- 39.Sakata H, Takayama H, Sharp R, Rubin J S, Merlino G, LaRochelle W J. Cell Growth Differ. 1996;7:1513–1523. [PubMed] [Google Scholar]
- 40.Smith G H, Sharp R, Kordon E C, Jhappan C, Merlino G. Am J Pathol. 1995;147:1081–1096. [PMC free article] [PubMed] [Google Scholar]
- 41.Saitoh K, Takahashi H, Sawada N, Parsons P G. J Pathol. 1994;174:191–199. doi: 10.1002/path.1711740308. [DOI] [PubMed] [Google Scholar]
- 42.Bottaro D P, Rubin J S, Faletto D L, Chan A M, Kmiecik T E, Vande Woude G F, Aaronson S A. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 43.Rong S, Oskarsson M, Faletto D, Tsarfaty I, Resau J H, Nakamura T, Rosen E, Hopkins R F, Vande Woude G F. Cell Growth Differ. 1995;4:563–569. [PubMed] [Google Scholar]
- 44.Ferracini R, Di Renzo M F, Scotlandi K, Baldini N, Olivero M, Lollini P, Cremona O, Campanacci M, Comoglio P M. Oncogene. 1995;10:739–749. [PubMed] [Google Scholar]
- 45.Cortner J, Vande Woude G F, Rong S. Experentia. 1995;74:89–121. doi: 10.1007/978-3-0348-9070-0_6. [DOI] [PubMed] [Google Scholar]
- 46.Biéche I, Champéme M H, Matifas F, Hacéne K, Callahan R, Lidereau R. Lancet. 1992;339:139–143. doi: 10.1016/0140-6736(92)90208-k. [DOI] [PubMed] [Google Scholar]
- 47.Liang T J, Reid A E, Xavier R, Cardiff R D, Wang T C. J Clin Invest. 1996;97:2872–2877. doi: 10.1172/JCI118744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boccaccio C, Gaudino G, Gambarotta G, Galimi F, Comoglio P M. J Biol Chem. 1994;269:12846–12851. [PubMed] [Google Scholar]
- 49.Takagi H, Sharp R, Hammerseister C, Goodrow T, Bradley M O, Fausto N, Merlino G. Cancer Res. 1992;52:5171–5177. [PubMed] [Google Scholar]
- 50.Amundadottir L T, Johnson M D, Merlino G, Smith G H, Dickson R B. Cell Growth Differ. 1995;6:737–748. [PubMed] [Google Scholar]
- 51.Andermarcher E, Surani M A, Gherardi E. Dev Genet. 1996;18:254–266. doi: 10.1002/(SICI)1520-6408(1996)18:3<254::AID-DVG6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 52.Niranjan B, Buluwela L, Yant J, Perusinghe N, Atherton A, Phippard D, Dale T, Gusterson B, Kamalati T. Development (Cambridge, UK) 1995;121:2897–2908. doi: 10.1242/dev.121.9.2897. [DOI] [PubMed] [Google Scholar]