Abstract
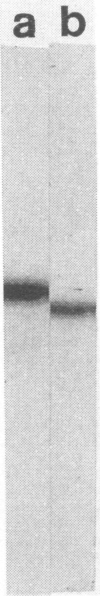
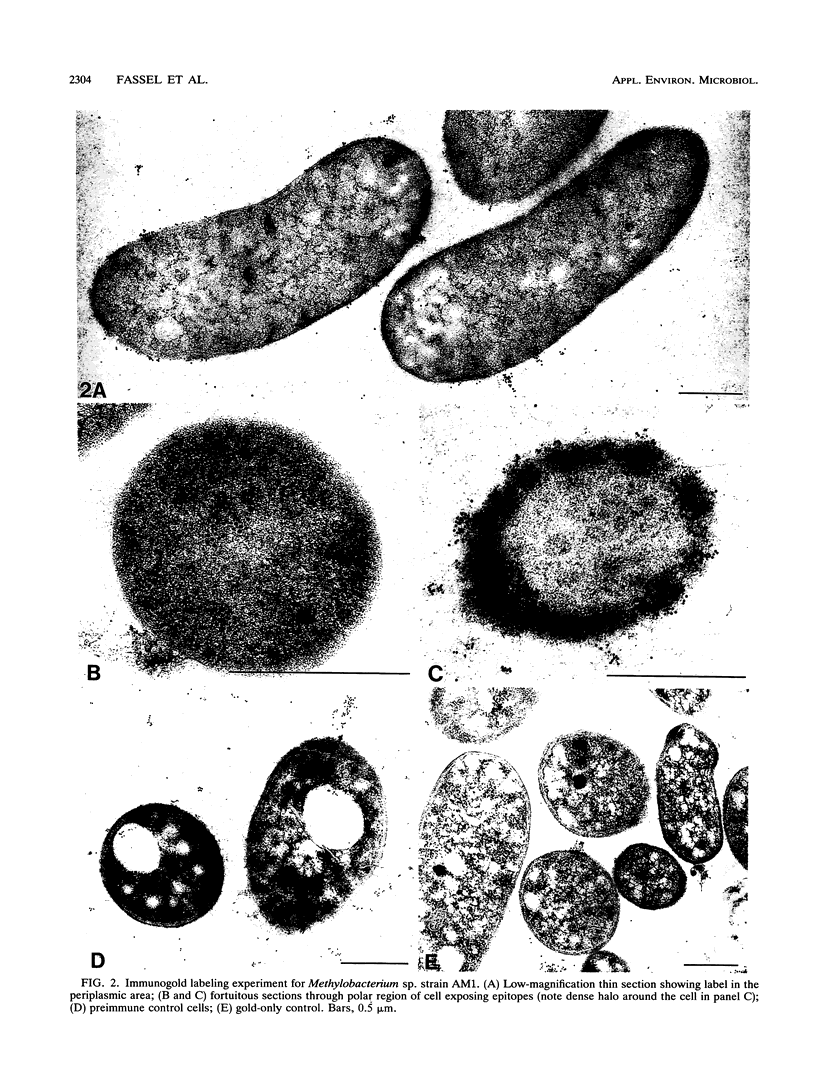
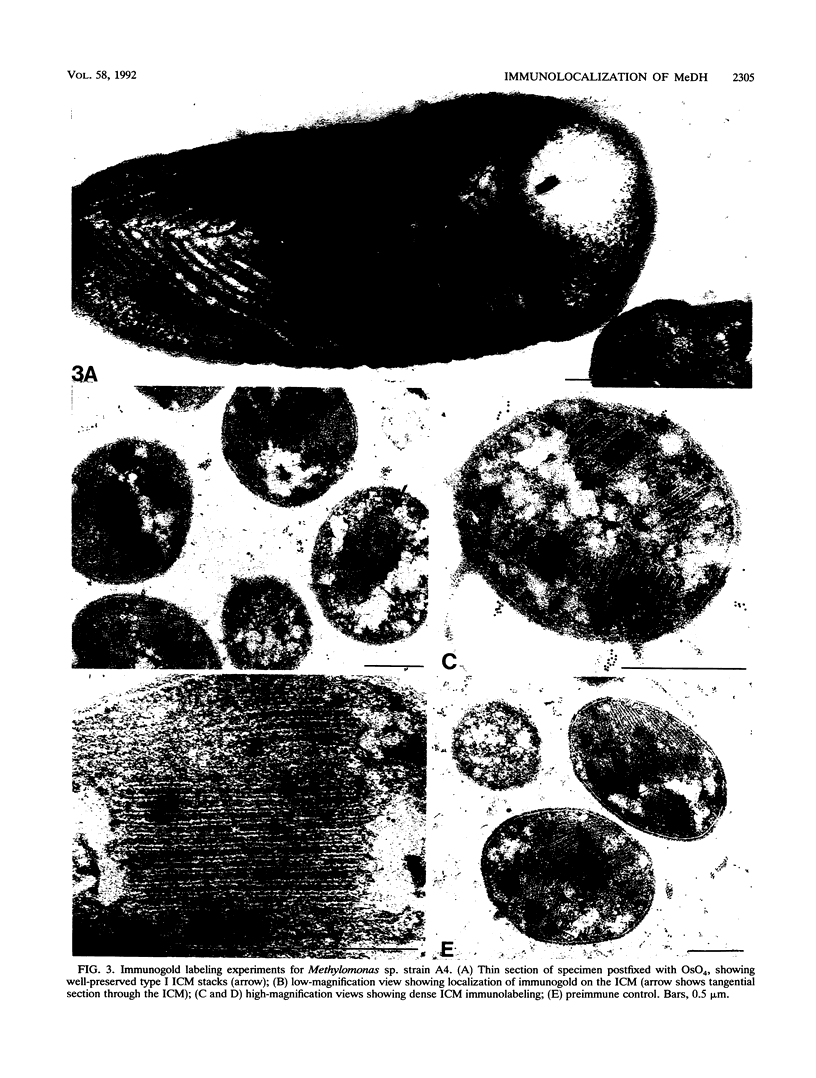
Antibodies to methanol dehydrogenase purified from Methylobacterium sp. strain AM1 and Methylomonas sp. strain A4 were raised. The antibody preparations were used in indirect immunogold labeling studies. With this approach, methanol dehydrogenase was found to be preferentially localized to the periplasmic region of the methylotroph Methylobacterium sp. strain AM1 and to the intracytoplasmic membrane of the methanotroph Methylomonas sp. strain A4. Antibody cross-reactivity to other methylotrophic bacteria was detected.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alefounder P. R., Ferguson S. J. A periplasmic location for methanol dehydrogenase from Paracoccus denitrificans: implications for proton pumping by cytochrome aa3. Biochem Biophys Res Commun. 1981 Feb 12;98(3):778–784. doi: 10.1016/0006-291x(81)91179-7. [DOI] [PubMed] [Google Scholar]
- Collins M. L., Buchholz L. A., Remsen C. C. Effect of Copper on Methylomonas albus BG8. Appl Environ Microbiol. 1991 Apr;57(4):1261–1264. doi: 10.1128/aem.57.4.1261-1264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook S. M., Treml S. B., Collins M. L. Immunocytochemical ultrastructural analysis of chromatophore membrane formation in Rhodospirillum rubrum. J Bacteriol. 1986 Jul;167(1):89–95. doi: 10.1128/jb.167.1.89-95.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W. E., Hazeu W. Observations on the fine structure of a methane-oxidizing bacterium. Antonie Van Leeuwenhoek. 1972;38(1):33–47. doi: 10.1007/BF02328075. [DOI] [PubMed] [Google Scholar]
- Ferenci T., Strom T., Quayle J. R. Oxidation of carbon monoxide and methane by Pseudomonas methanica. J Gen Microbiol. 1975 Nov;91(1):79–91. doi: 10.1099/00221287-91-1-79. [DOI] [PubMed] [Google Scholar]
- Ghosh R., Quayle J. R. Purification and properties of the methanol dehydrogenase from Methylophilus methylotrophus. Biochem J. 1981 Oct 1;199(1):245–250. doi: 10.1042/bj1990245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin E. J., Sorenson G. D., Dennett J. C., Cate C. C. Unlabeled antibody methods in electron microscopy: a comparison of single and multistep procedures using colloidal gold. J Histochem Cytochem. 1984 Aug;32(8):799–804. doi: 10.1177/32.8.6379035. [DOI] [PubMed] [Google Scholar]
- Kasprzak A. A., Steenkamp D. J. Localization of the major dehydrogenases in two methylotrophs by radiochemical labeling. J Bacteriol. 1983 Oct;156(1):348–353. doi: 10.1128/jb.156.1.348-353.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom M. E. Isolation and characterization of marine methanotrophs. Antonie Van Leeuwenhoek. 1988;54(3):189–199. doi: 10.1007/BF00443577. [DOI] [PubMed] [Google Scholar]
- Lynch M. J., Wopat A. E., O'connor M. L. Characterization of two new facultative methanotrophs. Appl Environ Microbiol. 1980 Aug;40(2):400–407. doi: 10.1128/aem.40.2.400-407.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin S. M., Hanson R. S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J Bacteriol. 1988 Oct;170(10):4739–4747. doi: 10.1128/jb.170.10.4739-4747.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Hou C. T., Felix A. Microbial oxidation of methane and methanol: crystallization of methanol dehydrogenase and properties of holo- and apomethanol dehydrogenase from Methylomonas methanica. J Bacteriol. 1978 Feb;133(2):641–649. doi: 10.1128/jb.133.2.641-649.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer K. P., Buchholz L. A., Lidstrom M. E., Remsen C. C. Separation of methanotrophic bacteria by using percoll and its application to isolation of mixed and pure cultures. Appl Environ Microbiol. 1991 Dec;57(12):3656–3659. doi: 10.1128/aem.57.12.3656-3659.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Tanaka K. Ultrastructure of intracytoplasmic membranes of Methanomonas margaritae cells grown under different conditions. Antonie Van Leeuwenhoek. 1980;46(1):15–25. doi: 10.1007/BF00422225. [DOI] [PubMed] [Google Scholar]
- Wadzinski A. M., Ribbons D. W. Oxidation of C1 compounds by particulate fractions from Methylococcus capsulatus: properties of methanol oxidase and methanol dehydrogenase. J Bacteriol. 1975 Jun;122(3):1364–1374. doi: 10.1128/jb.122.3.1364-1374.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H. J., Hanson R. S. Alcohol dehydrogenase from Methylobacterium organophilum. Appl Environ Microbiol. 1978 Jul;36(1):105–114. doi: 10.1128/aem.36.1.105-114.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]