Abstract
A gelatinous otolithic membrane (OM) couples a single calcified otolith to the sensory epithelium in the bluegill sunfish (Lepomis macrochirus) saccule, one of the otolithic organs in the inner ear. Though the OM is an integral part of the anatomic network of endorgan structures that result in vestibular function in the inner ear, the identity of the proteins that make up this sensory accessory membrane in teleosts, or in any vertebrate, is not fully known. Previously, we identified a cDNA from the sunfish saccular otolithic organ that encoded a new member of the collagen family of structural proteins. In this study, we examined biochemical features and the localization of the saccular collagen (SC) protein in vivo using polyclonal antisera that recognize the noncollagenous domains of the SC protein. The SC protein, in vivo, was identified as a 95-kDa glycoprotein in sunfish whole-saccule lysate and in homogenates of microdissected saccular OMs. Immunohistochemical analyses demonstrated that the SC protein was localized within one of the two distinct layers of the sunfish saccular OM. The SC protein was also detected within the cytoplasm of supporting cells at the edges of the saccular sensory epithelium, indicating that these cells are a primary site for the synthesis of this structural protein. Further studies of the organization of this matrix molecule in the OM may help clarify the role of this sensory accessory membrane in vestibular sensory function.
Keywords: vestibular, saccule, collagen, otolithic membrane, inner ear
The otolithic endorgans are part of the vestibular portion of the vertebrate inner ear and consist of a single or many calcified masses coupled to an underlying hair cell-containing sensory epithelium. A gelatinous otolithic/otoconial membrane is attached to both the otolith and the underlying sensory epithelium and serves to couple the calcified mass(es) to the receptor epithelium. Displacement of the overlying otolith/otoconial mass(es) relative to the underlying receptor epithelium results in the displacement of hair cell stereociliary bundles that are critical to the mechanoelectrical transduction process in the inner ear (1).
The teleost saccule is an otolithic organ involved in aspects of both vestibular (1) and acoustic function in certain fish (1, 2). The number of saccular sensory cells in certain teleosts (3) and cartilagenous fish (4) can be much larger than the number found in the human saccule (1). We took advantage of the large number of saccular hair cells found in certain teleosts to construct cDNA libraries in efforts to identify genes involved in vestibular function (5).
Differential screening of the teleost saccular cDNA libraries led to the identification of candidate saccule-specific cDNAs. One of these was found to encode a unique form of collagen that was termed saccular collagen (SC). The 2.0-kb SC transcript was highly abundant in saccular RNA and appears unique to the saccule because it was not detected in several other sunfish tissues (6). A similar domain organization and homology of a conserved region within the carboxyl-terminal noncollagenous domain identified the SC as a new member of the family of collagens that includes collagens type VIII and type X (7–9). Expression of the SC gene was localized by in situ hybridization to secretory supporting cells located around the periphery of the saccular macula (6). In teleosts, supporting cells such as these have been correlated with the developmental appearance of the otolithic membrane (OM) (10) suggesting that the SC may be a component of the teleost OM. To date, the molecular identity of the proteins that comprise the teleost OM remains unknown.
In this report, polyclonal SC-specific antisera were used to study the localization and biochemical features of the SC protein in vivo. Multiple antipeptide sera identify the SC protein as a 95-kDa glycoprotein in whole-saccule lysates and in homogenates of microdissected OM. The SC protein was detected in one discrete layer of the OM, establishing that the SC is a component of this sensory accessory membrane in the bluegill sunfish (Lepomis macrochirus) inner ear. We also detected the SC protein in a subset of saccular-supporting cells, suggesting that these cells may be involved in the postembryonic formation of one layer of the teleost OM. These studies demonstrate that the SC is located in part of the network of anatomic structures that facilitate vestibular function and provide additional information regarding the biochemical nature of the OM in the bluegill sunfish inner ear.
MATERIALS AND METHODS
Fish Care and Maintenance.
Bluegill sunfish (L. macrochirus) were obtained from Fender Fish Farm (Baltic, OH), and most were processed immediately upon arrival. The bluegill sunfish were between 5 and 6 inches in length and ≈200 fish were used during the course of these experiments. The sunfish were killed and the inner ear tissues removed and placed into lysis buffer for Western blot analysis (see below), or into 10% PBS-buffered formalin for 2 h before paraffin embedding and sectioning. The sunfish were maintained, and the tissues were obtained in accordance with the guidelines of the University of Pennsylvania’s Department of Laboratory Animal Management.
In Vitro Transcription/Translation Analysis of SC cDNA.
One microgram of a plasmid containing the full-length SC cDNA or the full-length luciferase cDNA was separately used as substrate for the coupled in vitro transcription/translation reaction (Promega) according to the manufacturer’s specifications. The 35S-labeled translation product(s) and control reactions were subjected to SDS/PAGE analysis in 7% polyacrylamide gels (11). The gels were fixed in 10% trichloroacetic acid solution for 30 min, dried, and subjected to standard autoradiography.
Generation of Anti-SC Sera.
Two peptides from the SC COOH noncollagenous (NC) domain, one termed C-NC1 corresponding to SC amino acids 350–364 (RKLRTRDSLYGQDID) and another termed C-NC2 corresponding to amino acids 376–388 (TDGDQVWLETLRD), and one peptide from the SC NH2-NC domain termed N-NC corresponding to SC amino acids 29–36 (APPGNTP) were synthesized by the Medical Center Protein Chemistry Facility of the University of Pennsylvania. blast searches (12) using the sequences of these peptides verified their SC specificity. These SC-specific peptides were coupled to chicken ovalbumin according to standard methodology (13) and used to immunize rabbits (Cocalico Biologicals, Reamstown, PA). The reactivity and specificity of the immune sera to the synthetic peptides was confirmed by analyzing the ability of dilutions of sera to detect the peptides immobilized on nitrocellulose. Detection of bound antibodies was performed as described for the Western blot analyses. In addition, 1 mg/ml of immunizing peptide, but not of irrelevant peptide, was able to inhibit detection of the 95-kDa band detected with 1:500 dilution of either the anti-N-NC or anti-C-NC1 sera (data not shown). In addition, the SC cDNA-derived transcription/translation product was recognized by the anti-C-NC1 sera (data not shown).
Tissue Lysate Preparation.
Bluegill sunfish sacculae were removed as described (6) and placed in 4°C lysis buffer (50 mM Tris·HCl, pH 7.5/150 mM NaCl/0.1% SDS) and subjected to mechanical homogenization using an Eppendorf pestel (Kontes Instruments). Tissue homogenates were boiled for 10 min and cleared of insoluble material by microcentrifugation at 5000 × g for 5 min. The recovered supernatant/tissue lysate was stored at −20°C. Equivalent volumes of the indicated tissue lysates/homogenates were analyzed in all lanes shown. Each lane of whole-saccule lysate contained lysate from approximately 1/10th of one saccule, and each lane of OM homogenate contained homogenate from approximately 1/20th of one OM.
Western Blot Analysis.
Aliquots of tissue lysate (approximately equivalent to 1/10 of saccular macula or 1/20 of an OM) were diluted with loading buffer, electrophoresed in 8% acrylamide gels, then transferred to nitrocellulose membrane (Schleicher & Schuell) at 2.5 mA/cm2 for 45 min. For spot test analysis, peptides were spotted onto nitrocellulose. The membranes were incubated in blocking solution (0.5× Blotto/5% goat serum in 1× PBS) for 1 h before incubation with the indicated sera diluted in 0.2× blocking solution for 2 h. Membranes were washed three times for 15 min in 1× PBS and incubated with 1:1000 goat anti-rabbit IgG conjugated to alkaline phosphatase (Boehringer Mannheim) diluted in 0.2× blocking solution. Membranes were washed again and equilibrated in developing solution (100 mM Tris·HCl/100 mM NaCl/50 mM MgCl2, pH 9.5) for 5 min. Membranes were then incubated with 470 nM each of X-phosphate and 4-nitrotetrazolium blue diluted in developing solution for 10 min. The reaction was quenched with stop solution (50 mM Tris·HCl, pH 7.2/5 mM EDTA).
Analysis of Bacterial Collagenase Sensitivity.
Aliquots of whole-saccule lysate or of microdissected OM homogenate were diluted in lysis buffer and CaCl2 was added to a final concentration of 10 mM. Bacterial collagenase was then added to a final concentration of 110 μg/μl. These and untreated samples were incubated at 37°C for 4 h. Reactions were stopped by addition of SDS/PAGE loading buffer and were assessed using Western blot analysis. Immune and preimmune sera were used at 1:500 dilutions.
Microdissection of Teleost OM for Western Blot Analysis.
During dissections, the OM remained attached to either the otolith or saccular epithelium and was recovered with a fine forceps and placed directly into lysis buffer and stored on ice. Harvested OMs were mechanically homogenized with Eppendorf pestels and then boiled for 10 min. Insoluble material was then pelleted by centrifugation at 10,000 × g for 10 min at room temperature. The supernatant was recovered and stored at −20°C until use.
Glycosylation Studies.
Saccular lysate and OM homogenate prepared as described above were supplemented with Triton X-100 to a final concentration of 1%. Aliquots of these preparations were incubated at 37°C for 4 h with 0.5 units of recombinant N-glycosidase-F (Boehringer Mannheim). An identical reaction was prepared without glycosidase as a negative control. Reactions were terminated by addition of 3× SDS/PAGE loading buffer before Western blot analysis.
Affinity Purification of SC-Specific Immunoglobulins.
Approximately 3 micromoles of the SC-specific peptide C-NC1 were coupled to cyanogen-bromide activated Sepharose (Sigma), yielding a column with a 3 ml bed volume and 3 μmol maximum immunoglobulin binding capacity. SC-peptide reactive immunoglobulins were adsorbed from the sera and eluted using low pH according to standard methodologies (13). The yield of C-NC1-specific immunoglobulins was 2.5 mg per 10 ml of production sera. The specificity of the C-NC1-specific immunoglobulins to the immunizing peptide and to the 95-kDa band, and lack of reactivity of the “flow-through” sera, was confirmed by spot test analysis and Western blot analysis (data not shown).
Immunohistochemical Analysis of Sunfish Saccular Maculae with Affinity-Purified Anti-SC Immunoglobulins.
Saccular maculae were removed and fixed for 1 h in 10% phosphate-buffered formalin and then rinsed extensively with cold 70% ethanol before embedding in paraffin. Sections 5 microns thick were deparaffinized by two successive 5-min immersions in xylene and were rehydrated through a descending series of ethanol into PBS and were stained with hematoxylin/eosin according to standard methodologies (14) or were used for immunohistochemistry. Sections were then covered in blocking solution (10% goat serum/0.5% nonfat dry milk in PBS) for 1 h before incubation with either 2.5 μg/ml of affinity-purified immunoglobulins or with preimmune sera diluted 1:500 in 0.5× blocking solution for 2 h, were washed three times for 10 min in PBS, and then incubated with goat anti-rabbit IgG Fab fragments coupled to alkaline phosphatase (Boehringer Mannheim) diluted 1:1000 in 0.5× blocking solution. Membranes were washed again then equilibrated in developing solution (100 mM Tris·HCl/100 mM NaCl/50 mM MgCl2, pH 9.5) for 5 min. SC localization was visualized by treatment with the X-phosphate and 4-nitroblue tetrazolium chloride (as described for Western blot analysis) for 2 h and then the stained sections were preserved with Glycergel (Sigma).
RESULTS
Estimation of Molecular Weight of SC Protein Using a Coupled in Vitro Transcription/Translation System.
The primary SC open reading frame is predicted to encode a 423-amino acid polypeptide with an estimated nonglycosylated molecular weight of ≈44.2 kDa. A cDNA containing the entire coding region of the SC gene was analyzed using a rabbit reticulocyte-derived coupled in vitro transcription/translation system (Promega). A single polypeptide of ≈55-kDa molecular weight was generated from the full-length SC cDNA whereas no product was generated when either the SC cDNA or the RNA polymerase was omitted from the reaction (Fig. 1). The observed molecular weight is larger than predicted and likely reflects the presence of the fibrillar domain of the SC protein that may slow its migration during SDS/PAGE analysis (15).
Figure 1.
In vitro transcription/translation analysis of the SC cDNA. One microgram of a plasmid containing the full-length SC cDNA or the full-length luciferase cDNA as a control were used as substrate for the rabbit reticulocyte-derived coupled in vitro transcription/translation reaction in the presence of [35S]methionine (Amersham). Control reactions in which cDNA template or RNA polymerase was omitted were also prepared and all the reaction products were resolved in 7% polyacrylamide denaturing gels as described. Lanes: NTP, no collagen template, with polymerase; LP, luciferase cDNA plus polymerase (62 kDa); CP, SC cDNA plus polymerase; NP, with collagen template but without polymerase. The arrowhead points to the in vitro transcription/translation product synthesized from the SC cDNA in lane CP.
Development of Polyclonal SC-Specific Antisera and Detection of the SC Protein in Whole-Saccule Lysate.
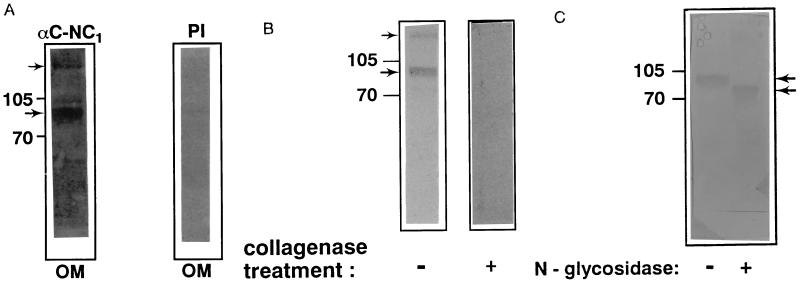
A schematic of the predicted domain organization of the SC protein and the location of the peptides used for antisera generation are shown in Fig. 2A. Two SC-specific peptides from the SC COOH-NC domain (one corresponding to SC amino acids 350–364 and one corresponding to amino acids 376–388) and one from the SC NH2-NC domain (corresponding to SC amino acids 29–36) were synthesized and used to generate polyclonal anti-SC sera (Cocalico Biologicals). These sera were reactive against the corresponding SC-specific peptide while the preimmune sera was not (Fig. 2B). Lysates were prepared from bluegill sunfish (L. macrochirus) whole-saccule and subjected to Western blot analysis. Both the anti-NH2-NC and the anti-COOH-NC domain sera detect a protein of ≈95-kDa molecular weight in whole-saccule lysates (Fig. 2C). There was little background in the preimmune sera at the 1:500 dilutions used in these experiments. A band of higher molecular weight was also observed with both of the anticarboxyl-terminal noncollagenous domain-specific sera, suggesting the possible existence of a covalently associated multimeric or heteromeric form of the SC protein in vivo. The higher molecular weight form was not detected with the anti-amino-terminal noncollagenous domain-specific sera suggesting that the epitopes recognized by this sera may no longer exposed as a consequence of the multimeric association. The specificity of detected SC protein in whole-saccule lysates was examined by several means. The immunizing peptide, but not an irrelevant peptide, was able to inhibit the detection of the 95-kDa band (data not shown). In addition, anti-SC protein C-NC1 sera was able to detect the 55-kDa in vitro transcription/translation product produced from the full-length SC cDNA (data not shown). In addition, both forms of the SC protein were sensitive to bacterial collagenase treatment (Fig. 3A).
Figure 2.
Generation of SC-specific antisera and detection of the SC protein in whole-saccule lysate. (A) Schematic of the predicted domain organization of the SC and the location of the immunizing peptides. The location of the amino and carboxy noncollagenous domains (N-NC and C-NC, respectively) are indicated. The SC consists of a single fibrillar, collagenous domain (indicated by the black bar) separating amino- and carboxyl-terminal noncollagenous domains (indicated by the black zig-zag pattern). The number of amino acids in each domain is indicated below that domain. The approximate location of the sequences of the SC-specific peptides are indicated by the boxed portions of the SC protein. C-NC1 and C-NC2, SC carboxyl-terminal noncollagenous domain peptides; N-NC, SC amino-terminal noncollagenous domain peptide. (B) The reactivity of sera to 1 μg of the indicated immunizing peptides and lack of reactivity to irrelevant peptides was confirmed by examining the ability of 10−2 and 10−3 dilutions of immune and preimmune sera to bind peptides immobilized on nitrocellulose. (C) Western blot analysis of whole sunfish saccule lysate with SC-specific antisera. In all panels, the position of the protein molecular weight markers (in kDa) is indicated with lines and the arrow(s) points at the detected protein(s). Each of the immune sera and the preimmune sera were diluted 1:500 for the analysis shown. Sera: anti-N-NC, SC amino-terminal noncollagenous domain peptide specific sera; anti-C-NC1 or anti C-NC2, SC carboxyl-terminal noncollagenous domain peptide-specific sera; or PI, preimmune sera.
Figure 3.
Western blot analysis of the SC protein in sunfish OM homogenate. In all panels, the position of the protein molecular weight markers (in kDa) is indicated with lines and the arrow(s) points at the detected protein(s). The immune sera and the preimmune sera were diluted 1:500 for the analysis shown. (A) Microdissected sunfish OM homogenate was probed with probed anti-C-NC1 or with preimmune (PI) sera. (B) Equivalent aliquots of the OM homogenate were incubated with or without bacterial collagenase (Sigma) for 4 h at 37°C before Western analysis using anti-C-NC1 antisera. (C) Equivalent aliquots of OM homogenate were treated with N-glycosidase-F (Boehringer Mannheim) according to the manufacturer’s specifications before Western analysis. The upper arrow points to the detected protein in the untreated OM homogenate, and the lower arrow points to the protein detected in the OM homogenate that had been treated with N-glycosidase.
Detection of the SC Protein in Microdissected OM Homogenate.
Homogenates from the OM were prepared and examined for the presence of the SC protein. The electrophoretic profile of the sunfish OM constituent in this homogenate (data not shown) approximately matches that of the trout OM (16). When probed with our anti-C-NC1 sera, the same two bands are detected in the OM homogenate as in the whole-saccule lysate (Fig. 3B). This demonstrates that both forms of the SC protein can be detected in the OM of the sunfish. No band was detected in several other fish tissue lysates including brain, eighth cranial nerve, gill, and semicircular canals using these same conditions (data not shown).
The SC Is Glycosylated in Vivo.
The two potential N-linked glycosylation sites in the SC protein (6) suggest that it is a glycoprotein. Therefore, OM homogenates were treated with N-glycosidase to assay for the presence of potential N-linked glycosylation of the SC protein. The increased mobility of the anti-SC sera detected bands indicates that ≈15 kDa of the 95-kDa mass was attributable to N-linked carbohydrates (Fig. 3C). As yet, we have not accounted for the additional 25 kDa of mass that, together with 15 kDa of N-linked glycosylation, account for the difference in mass between the 55-kDa SC in vitro transcription/translation product and the observed 95-kDa band detected by the anti-SC sera in the whole-saccule lysate.
Immunohistochemical Localization of the SC Protein Within the Teleost Saccule With Affinity Purified SC-Specific Immunoglobulins.
Sections of sunfish saccular maculae were incubated with purified C-NC1-specific immunoglobulins or with preimmune sera and the bound immunoglobulins were detected with a goat anti-rabbit IgG coupled to alkaline phosphatase. Affinity-purified SC protein-specific immunoglobulins stained the gelatinous, amorphous layer of the teleost OM (Fig. 4E) while the preimmune sera did not stain any regions of the saccular section examined (Fig. 4 C and D). The SC was uniformly distributed throughout the gelatinous layer of the OM (OMg) indicating that there was no regionally specific localization of the SC protein within this portion of the OM. The SC was not detected in the subcupular layer of OM (OMsc). Furthermore, the SC protein was also detected in the cytoplasm of the columnar supporting cells that reside at the margins of the saccular macula (arrowheads in Fig. 4F). This is consistent with the observed localization of the SC transcripts (6). However, only a subset of the marginal zone supporting cells contain the SC protein suggesting the possibility that discrete OMg synthesis sites are found within this population of cells.
Figure 4.
Immunohistochemical analysis of the localization of the SC protein within the teleost saccule. (A and B) Hematoxylin/eosin stained histologic view of the sections adjacent to those examined with the SC protein-specific immunoglobulins. The cross-sectional view of the saccular macula (sm) displays the hair cell and supporting cell layers within the sensory portion of the epithelium and is bounded by columnar, marginal zone (mz) supporting cells that populate the edges of the epithelium. The two layers of the OM are attached to one another but have been separated from the macula during the section preparation. The lower, subcupular layer (om-sc) and gelatinous layer (om-g) are seen here in a transverse section. The holes in the om-sc layer correspond to the area above each hair cell which is devoid of om-sc material resulting in a rounded clearing in this layer of the OM above each hair cell. (C and D) Saccular sections probed with preimmune sera at 1:500 dilution. (E and F) Saccular sections probed with 2.5 μg/ml of peptide-affinity purified anti-C-NC1 immunoglobulins. Arrowheads point to supporting cells at the edges of the sensory epithelium in which the SC protein could be detected. Bound SC-specific immunoglobulins were detected with goat anti-rabbit IgGs coupled to alkaline phosphatase and visualized using X-phosphate and 4-Nitro blue tetrazolium chloride development according to the manufacturer’s specifications (Boehringer Mannheim). (×40.)
DISCUSSION
In the vertebrate inner ear, sensory accessory membranes are associated with the hair cell-containing sensory epithelia. These specialized matrices are known as the cupula in the semicircular canals, the otolithic/otoconial membrane (OM) in otolithic organs, and the tectorial membrane in the auditory endorgan. These sensory accessory suprastructures are positioned over the corresponding inner ear sensory epithelia. In the otolithic organs, the OM couples the associated calcified mass (masses) to the underlying sensory epithelium. Displacements of the calcified mass (masses) relative to the sensory epithelium results in hair cell stereocilia displacement and subsequent hair cell depolarization or hyperpolarization (reviewed in refs. 17–20).
Each of the tectorial matrices is a discrete viscoelastic matrix comprised of a distinct mixture of glycoproteins collectively referred to as tectorins [constituents of the tectorial matrices (21)]. Owing to its small size, there have been no analytic electrophoretic studies of the constituents of isolated mammalian OMs. The only study of the constituents of an isolated vertebrate OM utilized the gelatinous OM of the trout saccule. The isolated gelatinous OM associated with the trout saccule contained at least six detectable glycoproteins (16). These studies indicated that novel matrix constituents may exist in the teleost OM.
In previous studies, we isolated and characterizated an inner ear-specific transcript in the bluegill sunfish saccule that encodes a new member (6) of the collagen superfamily of structural proteins (22, 23). The SC was determined to be a member of the subfamily of short chain collagens that includes collagens type VIII and type X (7–9). These collagens share a conserved domain organization (one short collagenous domain flanked by small globular domains) and a related subdomain in their respective carboxyl-terminal noncollagenous domains. The SC protein is distinguished from collagens type VIII and X by the smaller size of its collagenous domain and by its unique amino-terminal noncollagenous domain. In the postembryonic sunfish, transcripts encoding the SC protein were found in columnar supporting cells at the edges of the saccular sensory epithelium. During the embryonic development of the fish saccule, a comparable set of columnar cells appear responsible for the formation of the gelatinous OM (10), suggesting that the SC protein may be a constituent of the saccular OM.
In the present study, polyclonal antisera specific to the SC protein were used to examine its biochemical nature and localization in the sunfish inner ear. The SC protein has a predicted nonglycosylated molecular weight of 44.2 kDa and an observed mass of ≈55 kDa as assessed using in vitro transcription/translation analysis of full-length SC cDNA. Multiple polyclonal anti-SC peptide-specific sera each detected a protein exhibiting a mass of ≈95 kDa in whole-saccule lysates. This protein could not be detected with preimmune sera and was not observed in lysates from several other sunfish tissues. Sera that recognized each of the SC protein noncollagenous domains could detect the 95-kDa band indicating that portions of both the amino and carboxyl-terminal noncollagenous domains are retained in the mature SC protein in vivo. In addition, two of our sera detected a higher molecular weight protein that could indicate the presence of multimeric or heteromeric forms of this collagen. Within the saccule, the 95-kDa SC protein was specifically detected in homogenates of microdissected OMs establishing its presence in this membrane. Approximately 15 kDa of the observed 95-kDa SC protein’s mass was attributable to N-linked glycosylation in vivo. These data are consistent with the SC protein being the sunfish homologue of the 95-kDa protein biochemically identified in the trout OM (16).
We next examined the regional distribution of the SC protein within the teleost OM. The teleost OM consists of an upper gelantinous layer (OMg) and a lower filamentous, subcapular layer (OMsc) (24). Immunohistochemical analysis using affinity purified SC-reactive immunoglobulins indicated that the SC protein was uniformly dispersed throughout the gelatinous layer of the teleost OM. The SC protein was not detected in the subcupular layer of the OM. The SC protein was also detected within the cytoplasm of the marginal zone supporting cells (SCmz) that populate the periphery of the saccular epithelium, consistent with the localization of transcripts encoding the SC protein (6). These data further support the hypothesis that the SC protein is synthesized by the SCmz cells and secreted apically, presumably with other matrix constituents, to form the gelatinous layer of the postembryonic bluegill sunfish OM.
The inner ears of cartilagenous and bony fish continue to grow in fish that continue to grow throughout postembryonic life (3, 4). If the SC protein detected in the SCmz cells is secreted to become part of the OMg, then the addition of newly synthesized SC protein to the bluegill sunfish OMg may occur primarily at the edges of the saccular sensory epithelium via the SCmz cells. Though no SC protein was detected in supporting cells in the sensory portion of the epithelium, it is still possible that other sites of SC protein synthesis exist that were not detected in our immunohistochemical analysis. Studies of the development of the toadfish inner ear (10) have also implicated a comparable set of columnar supporting cells with the embryonic formation of the teleost OM. It is possible that the supporting cells involved in the embryonic formation of the teleost OMg (10) are functionally comparable to the supporting cells that synthesize SC protein in the postembryonic bluegill sunfish.
Though the precise supramolecular organization of the SC protein within the teleost OMg is not known, it may be related to that of collagens type VIII and X. When fully processed, these collagens retain the noncollagenous domains that flank their single, short collagenous domain. Monomers of these collagens assemble into covalently associated multimers, that then aggregate to form a three-dimensional, hexagonally arranged meshwork (25–27). The nodes of these meshworks are oligomerized noncollagenous globular domains and the internodal, fibrillar connections are oligomerized collagenous domains. A lattice similar to these was observed in the amphibian OM (28) suggesting there may be a correspondence between the SC protein and such a lattice, if such a lattice exists, in the teleost OM.
Some of our data are consistent with this possible correspondence. The amino- and carboxyl-terminal noncollagenous domains can be detected in the SC protein in the OM homogenate. By analogy to the collagens type VIII and X, the noncollagenous domains of the SC protein may aggregate to form the nodes of the meshwork. In addition, the higher molecular weight form of the SC protein detected in our analysis may be a multimer of the SC protein that could be the precursor subunit of such a lattice within the sunfish OMg. Furthermore, the uniform distribution pattern of the SC protein throughout the sunfish OMg is consistent with the distribution pattern that might be expected if such a lattice existed throughout the sunfish OMg.
These studies begin to define biochemical features of the SC protein in vivo and demonstrate the unique localization of this form of collagen in the bluegill sunfish saccule. At least one site (the SCmz cells) of the synthesis of the SC protein was identified. This site may be a primary site for the postembryonic formation of the gelatinous layer of the teleost OM. This is the first report in which the molecular identity of a protein of a teleost OM has been established. Further studies of the structural organization of the SC protein in the teleost OM may help clarify the role of this sensory accessory membrane in the mediation of vestibular mechanical sensitivity.
Acknowledgments
We thank Dr. James Saunders, Dr. Gail Massey, and Mr. Peter Stec for their assistance. This work was carried out with support from the Lucille P. Markey Charitable Trust and the Astral Corporation to M.I.G. S.I. was supported in part by a fellowship from Naito Foundation. J.G.D. was supported by National Institutes of Health Training Grant 1-T32-EY07131-03. This work was also supported by a grant to J.C.O from the National Institute on Deafness and Other Communication Disorders and the Pennsylvania Lion’s Hearing Research Foundation. This work was also supported by a grant to J.G.D. from the Deafness Research Foundation.
Footnotes
Abbreviations: SC, saccular collagen; OM, otolithic membrane; OMg, gelatinous layer of the OM; OMsc, subcupular layer of the OM.
References
- 1.Lewis E R, Leverenz E L, Bialek L S. The Vertebrate Inner Ear. Boca Raton, FL: CRC; 1985. [Google Scholar]
- 2.Popper A N, Fay R R. Brain Behav Evol. 1993;41:14–38. doi: 10.1159/000113821. [DOI] [PubMed] [Google Scholar]
- 3.Popper A N, Hoxter B. Hearing Res. 1984;15:133–142. doi: 10.1016/0378-5955(84)90044-3. [DOI] [PubMed] [Google Scholar]
- 4.Corwin J T. J Comp Physiol. 1981;142:379–390. [Google Scholar]
- 5.Davis J G, Oberholtzer J C, Burns F R, Lee A M, Saunders J, Eberwine J H, Greene M I. DNA Cell Biol. 1995;14:833–839. doi: 10.1089/dna.1995.14.833. [DOI] [PubMed] [Google Scholar]
- 6.Davis J G, Oberholtzer J C, Burns F R, Greene M I. Science. 1995;267:1031–1034. doi: 10.1126/science.7863331. [DOI] [PubMed] [Google Scholar]
- 7.LuValle P, Ninomiya Y, Rosenblum N D, Olsen B R. J Biol Chem. 1988;263:18378–18385. [PubMed] [Google Scholar]
- 8.Yamaguchi Y, Benya P D, van der Rest M, Ninomiya Y. J Biol Chem. 1989;264:16022–16029. [PubMed] [Google Scholar]
- 9.Muragaki Y, Jacenko O, Apte S, Mattei M-G, Ninomiya Y, Olsen B R. J Biol Chem. 1991;266:7721–7727. [PubMed] [Google Scholar]
- 10.Sokolowski B H A. Scanning Electron Microsc. 1986;4:1635–1648. [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Current Protocols in Immunology. New York: Wiley; 1995. [Google Scholar]
- 14.Luna L G, editor. Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology. New York: McGraw–Hill; 1968. [Google Scholar]
- 15.Butkowski R J, Noelken M E, Hudson B G. Methods Enzymol. 1982;82:410–423. [Google Scholar]
- 16.Khan K M, Drescher D G. Hearing Res. 1990;43:149–158. doi: 10.1016/0378-5955(90)90224-d. [DOI] [PubMed] [Google Scholar]
- 17.Hudspeth A J. Annu Rev Neurosci. 1983;6:187–215. doi: 10.1146/annurev.ne.06.030183.001155. [DOI] [PubMed] [Google Scholar]
- 18.Hudspeth A J. Nature (London) 1989;341:397–404. doi: 10.1038/341397a0. [DOI] [PubMed] [Google Scholar]
- 19.Corwin J T, Warchol M E. Annu Rev Neurosci. 1991;14:301–333. doi: 10.1146/annurev.ne.14.030191.001505. [DOI] [PubMed] [Google Scholar]
- 20.Pickles J O, Corey D P. Trends Neurosci. 1992;15:254–260. doi: 10.1016/0166-2236(92)90066-h. [DOI] [PubMed] [Google Scholar]
- 21.Killick R, Legan P K, Malenczak C, Richardson G P. J Cell Biol. 1995;129:535–547. doi: 10.1083/jcb.129.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgeson R E. Annu Rev Cell Biol. 1988;4:551–577. doi: 10.1146/annurev.cb.04.110188.003003. [DOI] [PubMed] [Google Scholar]
- 23.van der Rest M, Bruckner P. Curr Opin Struct Biol. 1993;3:430–436. [Google Scholar]
- 24.Dunkelberger D G, Dean J M, Watabe N. J Morphol. 1980;163:367–377. doi: 10.1002/jmor.1051630309. [DOI] [PubMed] [Google Scholar]
- 25.Sawada H, Konomi H, Nagai Y. Eur J Cell Biol. 1984;35:226–234. [PubMed] [Google Scholar]
- 26.Sawada H, Konomi H, Hirosawa K. J Cell Biol. 1990;110:219–227. doi: 10.1083/jcb.110.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwan A P L, Cummings C E, Chapman J A, Grant M E. J Cell Biol. 1991;114:597–604. doi: 10.1083/jcb.114.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kachar B, Parakkal M, Fex J. Hearing Res. 1990;45:179–190. doi: 10.1016/0378-5955(90)90119-a. [DOI] [PubMed] [Google Scholar]