Abstract
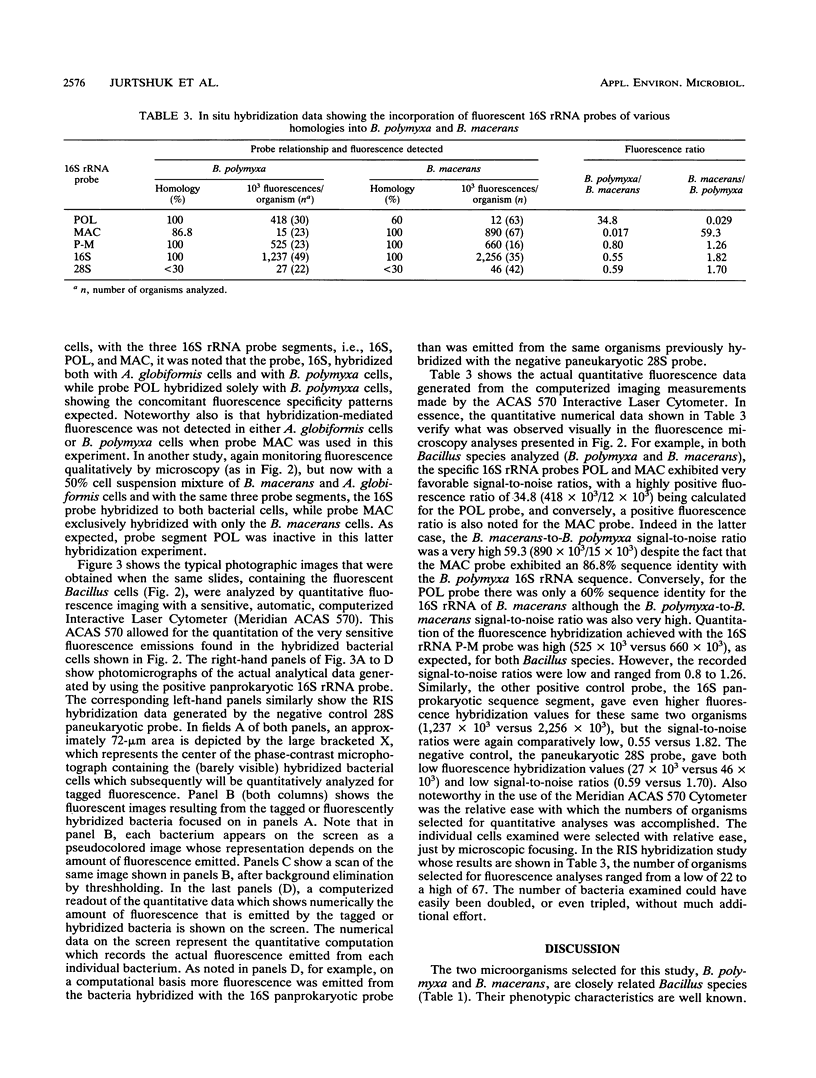
A rapid, sensitive, inexpensive in situ hybridization technique, using 30-mer 16S rRNA probes, can specifically differentiate two closely related Bacillus spp., B. polymyxa and B. macerans. The 16S rRNA probes were labeled with a rhodamine derivative (Texas Red), and quantitative fluorescence measurements were made on individual bacterial cells. The microscopic fields analyzed were selected by phase-contrast microscopy, and the fluorescence imaging analyses were performed on 16 to 67 individual cells. The labeled 16S rRNA probe, POL, whose sequence was a 100% match with B. polymyxa 16S rRNA but only a 60% match with B. macerans 16S rRNA, gave quantitative fluorescence ratio measurements that were 34.8-fold higher for B. polymyxa cells than for B. macerans cells. Conversely, the labeled probe, MAC, which matched B. polymyxa 16S rRNA in 86.6% of its positions and B. macerans 16S rRNA in 100% of its positions, gave quantitative fluorescence measurements that were 59.3-fold higher in B. macerans cells than in B. polymyxa cells. Control probes, whose 16S rRNA sequence segment (P-M) was present in both B. polymyxa and B. macerans as well as a panprokaryotic probe (16S), having a 100% match with all known bacteria, hybridized equally well with both organisms. These latter hybridizations generated very high fluorescence signals, but their comparative fluorescence ratios (the differences between two organisms) were low. The control paneukaryotic probe (28S), which had less than 30% identity for both B. macerans and B. polymyxa, did not hybridize with either organism.
Full text
PDF
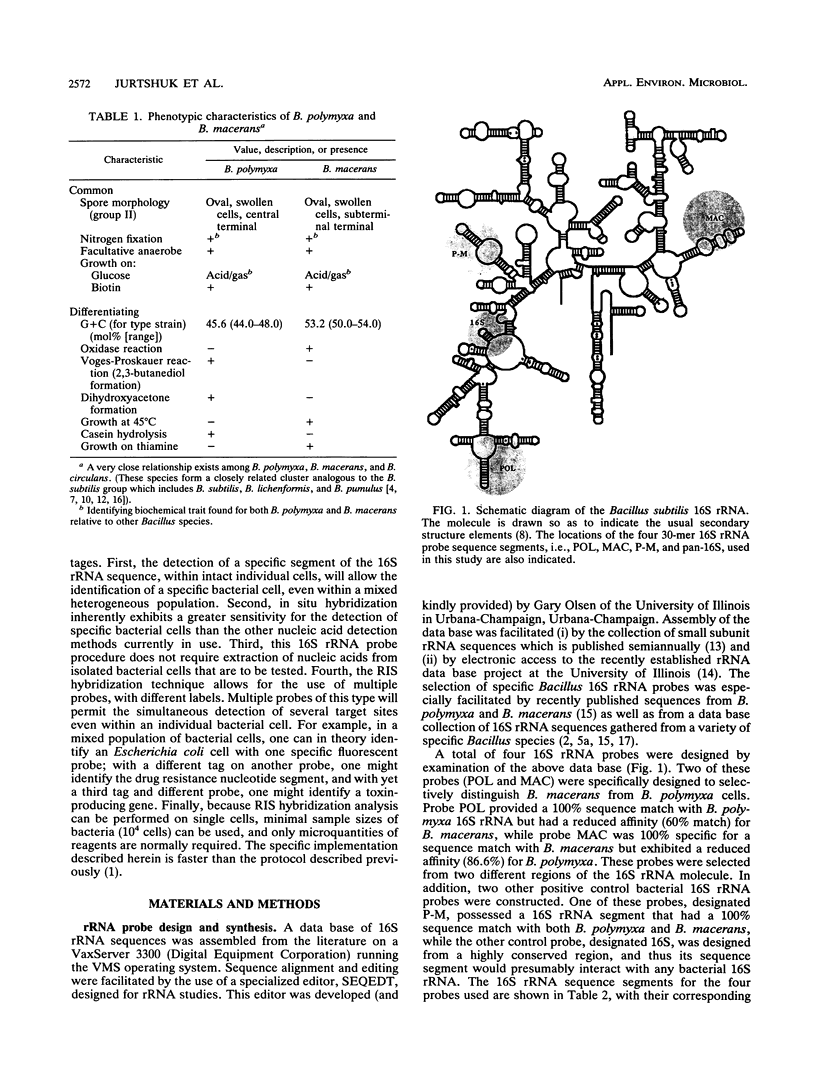





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash C., Farrow J. A., Dorsch M., Stackebrandt E., Collins M. D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991 Jul;41(3):343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Gonzalez I. L., Gorski J. L., Campen T. J., Dorney D. J., Erickson J. M., Sylvester J. E., Schmickel R. D. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Jones C. L., Sprecher C. J., Schumm J. W. An oligonucleotide probe to assay lysis and DNA hybridization of a diverse set of bacteria. Anal Biochem. 1989 Aug 15;181(1):23–27. doi: 10.1016/0003-2697(89)90388-6. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Larsen N., Woese C. R. The ribosomal RNA database project. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2017–2021. doi: 10.1093/nar/19.suppl.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler D., Ludwig W., Schleifer K. H., Lin C., McGill T. J., Wisotzkey J. D., Jurtshuk P., Jr, Fox G. E. Phylogenetic diversity in the genus Bacillus as seen by 16S rRNA sequencing studies. Syst Appl Microbiol. 1991;14(3):266–269. doi: 10.1016/S0723-2020(11)80379-6. [DOI] [PubMed] [Google Scholar]
- Wisotzkey J. D., Jurtshuk P., Jr, Fox G. E. PCR amplification of 16S rDNA from lyophilized cell cultures facilitates studies in molecular systematics. Curr Microbiol. 1990;21:325–327. doi: 10.1007/BF02092099. [DOI] [PubMed] [Google Scholar]




