Abstract
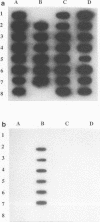
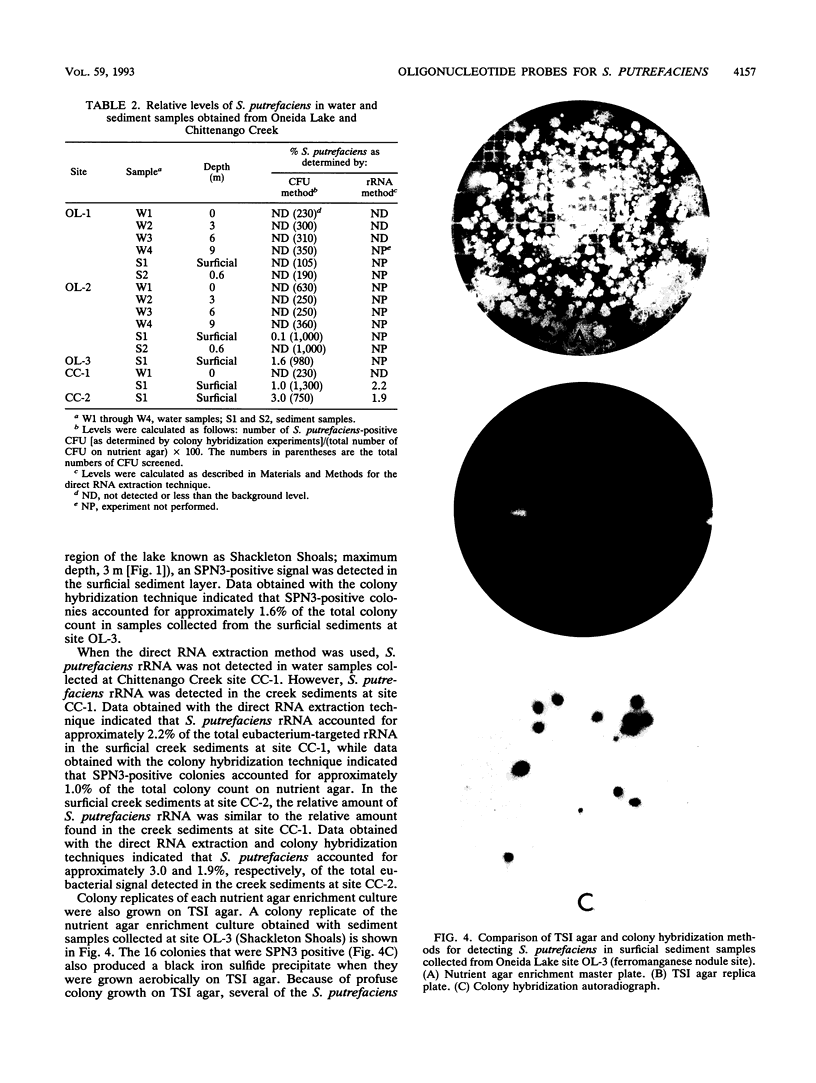
A 16S rRNA-targeted oligonucleotide probe specific for the iron (Fe3+)- and manganese (Mn4+)-reducing bacterium Shewanella putrefaciens was constructed and tested in both laboratory- and field-based hybridization experiments. The radioactively labeled probe was used to detect S. putrefaciens in field samples collected from the water column and sediments of Oneida Lake in New York and its major southern tributary, Chittenango Creek. S. putrefaciens was quantified by (i) hybridization of the probe to bulk RNA extracted from field samples and normalization of the S. putrefaciens-specific rRNA to total eubacterial rRNA, (ii) a colony-based probe hybridization assay, and (iii) a colony-based biochemical assay which detected the formation of iron sulfide precipitates on triple-sugar iron agar. The results of field applications indicated that the three detection methods were comparable in sensitivity for detecting S. putrefaciens in water column and sediment samples. S. putrefaciens rRNA was detected in the surficial layers of the lake and creek sediments, but the levels of S. putrefaciens rRNA were below the detection limits in the lake and creek water samples. The highest concentrations of S. putrefaciens rRNA, corresponding to approximately 2% of the total eubacterial rRNA, were detected in the surficial sediments of Chittenango Creek and at a midlake site where the Oneida Lake floor is covered by a high concentration of ferromanganese nodules. This finding supports the hypothesis that metal-reducing bacteria such as S. putrefaciens are important components in the overall biogeochemical cycling of iron, manganese and other elements in seasonally anoxic freshwater basins.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Stromley J., Devereux R., Key R., Stahl D. A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992 Feb;58(2):614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Arnold R. G., DiChristina T. J., Hoffmann M. R. Inhibitor studies of dissimilative Fe(III) reduction by Pseudomonas sp. strain 200 ("Pseudomonas ferrireductans") Appl Environ Microbiol. 1986 Aug;52(2):281–289. doi: 10.1128/aem.52.2.281-289.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E. L., Clark M. A. Tetrathionate reduction and production of hydrogen sulfide from thiosulfate. Microbiol Rev. 1987 Jun;51(2):192–205. doi: 10.1128/mr.51.2.192-205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Deming J. W., Hada H., Colwell R. R., Luehrsen K. R., Fox G. E. The ribonucleotide sequence of 5s rRNA from two strains of deep-sea barophilic bacteria. J Gen Microbiol. 1984 Aug;130(8):1911–1920. doi: 10.1099/00221287-130-8-1911. [DOI] [PubMed] [Google Scholar]
- DiChristina T. J. Effects of nitrate and nitrite on dissimilatory iron reduction by Shewanella putrefaciens 200. J Bacteriol. 1992 Mar;174(6):1891–1896. doi: 10.1128/jb.174.6.1891-1896.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E. L., Amaral L. A., Caron D. A., DeLong E. F. Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl Environ Microbiol. 1993 May;59(5):1647–1655. doi: 10.1128/aem.59.5.1647-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev. 1991 Jun;55(2):259–287. doi: 10.1128/mr.55.2.259-287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Giovannoni S. J., White D. C., Champine J. E., Phillips E. J., Gorby Y. A., Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159(4):336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- Lovley D. R., Phillips E. J. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988 Jun;54(6):1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C. R., Nealson K. H. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988 Jun 3;240(4857):1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- Myers C. R., Nealson K. H. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol. 1990 Nov;172(11):6232–6238. doi: 10.1128/jb.172.11.6232-6238.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H., Myers C. R. Microbial reduction of manganese and iron: new approaches to carbon cycling. Appl Environ Microbiol. 1992 Feb;58(2):439–443. doi: 10.1128/aem.58.2.439-443.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuekwe C. O., Westlake D. W., Cook F. D. Effect of nitrate on reduction of ferric iron by a bacterium isolated from crude oil. Can J Microbiol. 1981 Jul;27(7):692–697. doi: 10.1139/m81-107. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Overbeek R., Larsen N., Marsh T. L., McCaughey M. J., Maciukenas M. A., Kuan W. M., Macke T. J., Xing Y., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1992 May 11;20 (Suppl):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L. K., Ballard G., Stahl D. A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993 May;59(5):1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. M., DeLong E. F., Pace N. R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991 Jul;173(14):4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayanos A. A. Evolutional and ecological implications of the properties of deep-sea barophilic bacteria. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9542–9546. doi: 10.1073/pnas.83.24.9542. [DOI] [PMC free article] [PubMed] [Google Scholar]