Abstract
Vesicle traffic between the endoplasmic reticulum and the Golgi apparatus in mammals requires the small GTP-binding protein Rab2, but Saccharomyces cerevisiae appears not to have a Rab2 homolog. Here it is shown that the higher plant, Arabidopsis thaliana, contains a gene, At-RAB2, whose predicted product shares 79% identity with human Rab2 protein. Transgenic plants containing fusions between β-glucuronidase and sequences upstream of At-RAB2 demonstrated histochemical staining predominantly in maturing pollen and rapidly growing organs of germinating seedlings. β-glucuronidase activity in pollen is first detectable at microspore mitosis and increases thereafter. In this respect, the promoter of At-RAB2 behaves like those of class II pollen-specific genes, whose products are often required after germination for pollen tube growth. Seedling germination and pollen tube growth are notable for their unusually high rates of cell wall and membrane biosynthesis. These results are consistent with a role for At-RAB2 in secretory activity.
Transport of molecules destined for the vacuole, plasma membrane, endoplasmic reticulum (ER), Golgi, and cell wall is thought to occur largely in membrane vesicles of the exocytotic and endocytotic pathways. In plants, such transport pathways are important for storage-protein accumulation, for cell growth and differentiation, for secretion of protein and polysaccharide components of the cell wall and cell plate, and for morphogenesis, which depends on spatial and quantitative control of cell expansion.
From studies of yeasts and mammalian cells it has become evident that many organelles of the intracellular transport pathways have at least one type of small GTP-binding protein on their cytoplasmic face. These proteins are all members of the diverse Ras superfamily (1) and include ADP ribosylation factors, SAR1, and members of the YPT/Rab subfamily (2, 3). The YPT/Rab family can be subdivided into numerous structurally and functionally related subclasses, each of which acts at a particular transport step. It has been proposed that YPT/Rab family proteins are involved in regulating the fusion of transported vesicles with their appropriate target membranes. YPT/Rab proteins may regulate the interaction of v-SNAREs, molecular tags on transport vesicles, with t-SNAREs, docking proteins on target membranes (2–5).
Relatively little is known about how the plant endomembrane system performs its requisite biosynthetic and transport functions, or how these processes influence the growth, differentiation, and activity of the various plant cell types. To begin to address these questions we and others have isolated genes of the YPT/rab family from plants (reviewed in refs. 6–9). In mammalian cells Rab2 has been immunolocalized to a membrane compartment between the ER and Golgi and found to copurify with a marker for the Intermediate Compartment, which may be responsible for retrieving resident ER proteins that have escaped the ER (10). Furthermore, dominant–negative Rab2 mutants inhibit membrane traffic between mammalian ER and Golgi (11). Although many mammalian Rab proteins have homologs in other eukaryotes, it seems that no Rab2 homolog is present in the yeast genome (12). Here we report that a Rab2 homolog is present in the higher plant, Arabidopsis thaliana. We show that the Arabidopsis rab2 homolog is highly expressed in cells with high demand for endomembrane biogenesis, consistent with a function in ER to Golgi transport.
MATERIALS AND METHODS
General Cloning Techniques.
These cloning techniques were performed essentially as described in refs. 13 and 14. Genomic and cDNA libraries cloned in EMBL4 and Lambda-GEM-2 were screened according to ref. 7. A VAX-VMS computer with the Genetics Computer Group software package was used for sequence analysis.
RNA Isolation and Hybridization.
All plants used for RNA isolation were greenhouse grown. The Arabidopsis suspension culture was kindly provided by Mike May (Department of Plant Sciences, University of Oxford). Seedlings from a seed batch that showed synchronous germination were sown on several 9-cm filter paper discs soaked in 2 ml water and placed in Petri dishes, which were sealed with parafilm and incubated vertically at 26°C with 16 hr light (see Fig. 6C). Poly(A)-enriched RNA was isolated using paramagnetic beads (Dynal, Hamburg, Germany) according to the manufacturer’s recommendations, except that proteinase K was added at 200 μg/ml to the extraction buffer, and the RNA was eluted from the beads in formamide. Equal amounts of RNA from each sample (≈1 μg), estimated by absorbance at 260 nm, were separated on a denaturing formaldehyde gel (0.67% formaldehyde) and blotted onto Hybond-N+ (Amersham).
F1 seeds from an Arabidopsis plant heterozygous for the male sterile mutation ms1 were obtained from the Nottingham Arabidopsis Stock Center (Cat. No. NW75) (see Fig. 6B). The eIF4A probe used for this blot was isolated as a 390-bp XbaI–BamHI fragment from a plasmid provided by Pauline Bariola and Pam Green (Michigan State University–Department of Energy, East Lansing).
A 3′ At-RAB2 gene-specific probe was generated by first amplifying almost the entire cDNA with BamHI sites at each end using the primers 5′-GCGGATCCAGATTCTCTTCTCTTCTCG-3′ and 5′-GGATCCGCAAACACATCTTAATTTT-3′. A 337-bp PvuII–BamHI fragment was isolated as a probe. This fragment was labeled using [α-32P]dATP because it contains 62% dA and dT.
Construction of β-Glucuronidase (GUS) Fusions and Plant Transformation.
A 4.7-kb EcoRI fragment from the genomic phage λ6 was treated with Klenow fragment to remove the single stranded overhangs and cloned in the SmaI site of pBI101.3 (15). Two independent but apparently identical clones, p47gus1 and p47gus9, were used to transform Agrobacterium tumefaciens strain GV3101::pMP90 (16). Two A. tumefaciens transformants were selected for each of the duplicate plasmids p47gus1 and p47gus9 and used to transform Arabidopsis root explants essentially according to ref. 17, with modifications explained in ref. 18.
Analysis of GUS Activity.
To analyze GUS expression, plants were grown under sterile conditions in the presence of 50 μg/ml kanamycin until the resistant phenotype became clear, whereupon they were transferred to soil and grown to maturity in the greenhouse. Samples for histochemical analysis were vacuum infiltrated in 50 mM sodium phosphate buffer (pH 7.0), with 0.005% Tween-80 and 0.3% formaldehyde and fixation was continued for 30 min at room temperature. The tissue was washed three times in 50 mM sodium phosphate (pH 7.0) and stained for between 30 min and 18 h in 50 mM sodium phosphate (pH 7.0), 10 mM EDTA, 0.01% Tween-80, and 0.5 mg/ml X-gluc (Biosynth, Basel), which was dissolved initially at 100 mg/ml in dimethylformamide. After staining, the samples were washed in 50 mM sodium phosphate buffer (pH 7.0) and fixed for 15 min at room temperature in 20% ethanol, 5% acetic acid, and 5% formaldehyde (vol/vol). Chlorophyll-containing tissues were cleared in a series of ethanol:water mixtures up to 80% ethanol, in which samples were stored.
GUS assays were performed as described (15) on tissues collected from plants grown under sterile conditions in cotton–wool stoppered glass tubes. Protein determination on 5-μl aliquots of cleared tissue extracts was performed with Bio-Rad Protein Assay Reagent as recommended. Bovine serum albumin dissolved in assay buffer was used as a standard. Fluorescence of 4-methyl-umbelliferone at 455 nm was measured with a Perkin–Elmer fluorometer.
RESULTS
Identification of a Rab2 Homolog in Arabidopsis.
We previously screened a maize cDNA library for clones that show homology to the mammalian Rab family, and have reported the isolation of two Rab1 homologs (7). While analyzing other putative Rab homologs, we identified a third cDNA clone that was found to be similar in sequence to mammalian Rab2 (unpublished data). We used this clone to isolate a Rab2 homolog, At-RAB2, from a genomic DNA library of the model plant species A. thaliana.
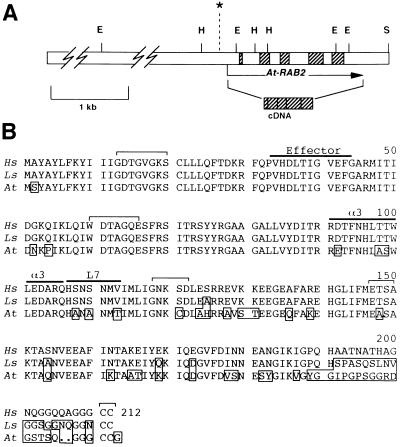
At-RAB2 was found to comprise five exons (Fig. 1) flanked by consensus intron–exon borders (20), predicting an open reading frame of 633 bp. Southern blot hybridization to restriction digests of Arabidopsis genomic DNA confirmed the genomic organization determined from the cloned DNA (not shown). The sequence of the open reading frame was confirmed by isolation and sequencing of a cDNA clone (Fig. 2). This also confirmed that At-RAB2 is transcribed (see below). Comparison of the deduced protein product, At-Rab2, with known Ypt/Rab subclasses confirmed that it showed most similarity to the Rab2 subclass (79% identity to canine Rab2; calculated from alignments generated by the fasta program of the Genetics Computer Group package (21).
Figure 1.
(A) Scale drawing of the genomic region surrounding the A. thaliana Rab2 locus. The open bar indicates the cloned genomic sequence. The exons are indicated by hatched boxes. The arrow indicates the direction of transcription, and its ends mark the ends of the available cDNA sequence. The asterisk indicates the position of the putative TATA box. E, EcoRI; H, HindIII; S, SalI. (B) The sequence of At-Rab2 (At) and Rab2 homologs from humans (Hs) and the gastropod Lymnaea stagnalis (Ls; ref. 19) were aligned and are shown. The conserved sequences involved in nucleotide binding and hydrolysis (GxxGVGKS, WDTAGQE, GNKxD, and ExSA) are marked by brackets, as are the conserved carboxyl-terminal cysteine residues; over-lining indicates the effector region (corresponding to residues 31–41 of Ha-Ras) and α-helix3 (α3) and loop7 (L7), which contain subfamily specific sequences implicated in determining subfamily specificity. Residues that differ from the mammalian Rab2 sequences are boxed.
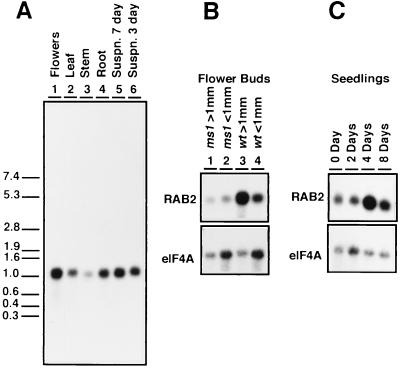
Figure 2.
(A) At-RAB2 transcripts in poly(A)-enriched RNA from the major organs of mature plants, and dividing (3 day) and stationary phase (7 day) suspension culture cells. The lengths (in kb) and positions of the RNA molecular weight markers are indicated on the left. (B) Flower buds estimated to be greater or less than 1 mm in length were collected separately from both wild type (wt) and male sterile ms1 mutant plants. Fully open flowers were not included in the >1-mm sample. As a control, the blot was reprobed with an Arabidopsis translation initiation factor clone, eIF4A, showing that expression of this gene was similar in wild-type and ms1 mutant buds. (C) RNA was isolated from unimbibed seeds (0 day) and 2, 4, and 8 days after imbibition. The blot was reprobed with the eIF4A clone as a control.
Expression of At-RAB2.
To begin to investigate the function of At-RAB2 we wished first to determine in which organs and tissues it was expressed. Fig. 2A shows that a single hybridizing band was detectable on RNA gel blots of poly(A)+-enriched RNA from the major organs and suspension cultured cells. This could have resulted from expression of the gene in most cells at a relatively low level, or from expression in a subpopulation of cells at a relatively high level, or both. To investigate these possibilities, the promoter activity of the upstream sequences was analyzed using promoter–GUS fusions. A 4.7-kb genomic EcoRI fragment that lies upstream of the At-RAB2 coding region (Fig. 1) was used to construct transcriptional fusions to the GUS reporter gene (15). This fragment includes a putative TATA box (see Fig. 1) and the first 81 bp of the 5′ untranslated sequence of the At-RAB2 cDNA. The At-RAB2–GUS fusion was transferred into Arabidopsis (R0 generation) and the progeny of transgenic R2 plants were analyzed for GUS expression.
Histochemical staining for GUS activity in organs from mature plants revealed a rather restricted distribution as shown in Table 1. In the majority of lines GUS activity was restricted to the anthers. Sporophytic tissues in the anther were not visibly stained (Fig. 3 a and b), but pollen washed out of the anthers stained well (Fig. 3e). In the lines that showed the highest GUS activities (47–1; 3.5–2 and 47–1; 3.1–2, Table 1) staining was strongest in the pollen but was repeatedly observed in the vascular tissues of most vegetative and floral organs examined. These data suggested that At-RAB2–GUS was expressed preferentially in pollen, but was active to some extent in many other cell types, which were visibly stained only if individual transformants express sufficient GUS activity. This conclusion was supported by direct measurement of biochemical activities (Fig. 4), which demonstrated that although stamens showed the most activity, GUS activity was above background in all tissues assayed.
Table 1.
Histochemical staining of At-RAB2–GUS transgenics
| Plant | Leaf | Stem | Sepal | Petal | Anther | Filament | Ovary | Silique | Root |
|---|---|---|---|---|---|---|---|---|---|
| 35S | +++ | +++ | ++ | ++ | + | + | +++ | ++ | +++ |
| Control | − | − | − | − | − | − | − | − | − |
| 47-1; 4.0 −2 | − | − | − | − | − | − | − | − | ND |
| 47-1; 4.0 −3 | − | − | − | − | + | − | − | − | ND |
| 47-9; 2.1 −3 | − | − | − | − | + | − | − | − | − |
| 47-9; 2.1 −2 | − | − | − | − | ++ | − | − | − | − |
| 47-9; 2.3 −1 | − | − | − | − | ++ | − | − | − | − |
| 47-9; 2.3 −2 | − | − | − | − | ++ | − | − | − | − |
| 47-1; 3.12 −1 | − | − | − | − | ++ | − | − | − | ND |
| 47-1; 2.2 −1 | − | − | − | − | ++ | − | − | − | − |
| 47-1; 2.2 −2 | − | − | − | − | ++ | − | − | − | − |
| 47-9; 2.5 −2 | − | − | − | − | +++ | − | − | − | − |
| 47-9; 2.5 −1 | − | − | + | + | +++ | + | − | − | − |
| 47-1; 3.5 −1 | − | (+) | (+) | (+) | +++ | − | − | − | ND |
| 47-1; 3.5 −2 | − | (+) | + | + | +++ | + | − | − | ND |
| 47-1; 3.1 −3 | − | − | − | − | ++ | − | − | − | ND |
| 47-1; 3.1 −1 | − | (+) | (+) | (+) | ++ | − | − | − | − |
| 47-1; 3.1 −2 | (+) | (+) | ++ | ++ | +++ | + | + | + | + |
Plants from eight transformed lines were dissected into their constituent parts and samples of each organ (intact and sectioned to allow substrate access) were stained for GUS activity. The staining was scored as follows: −, tissue unstained; +, ++, +++, tissue stained reflecting an estimate of intensity; (+), cases which could not be unambiguously assigned − or +. Transgenic lines are arranged from top to bottom in a rough order of staining intensity. Untransformed plants and a plant transgenic for a CaMV35S–GUS fusion (35S) were used as negative and positive controls, respectively. Other transgenic plants are numbered as follows: the first five digits define the initial transformant, from which seeds were obtained; from these seeds kanamycin resistant f1 siblings (numbered −1, −2, or −3) were grown to generate seed populations from which the f2 plants analyzed here were grown. ND, not determined.
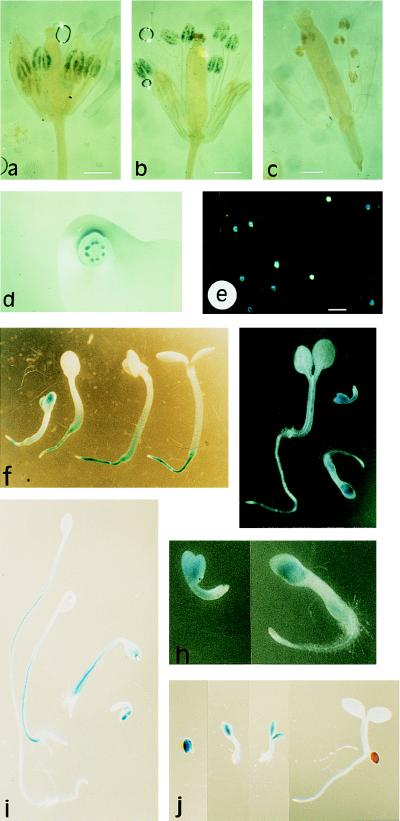
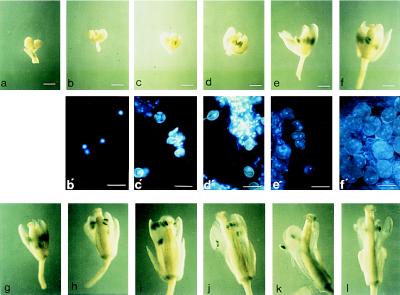
Figure 3.
Histochemical localization of GUS activity in plants transgenic for the At-RAB2–GUS fusion. (a) Young flower of line 47–9; 2.1 showing staining of the developing pollen grains in the four most developmentally advanced anthers. The two less advanced anthers lying beneath these four show no staining. (b) Older flower, post anthesis, in which pollen grains in all six anthers are stained. Pollen can be seen on the stigma. (c) Flower from an untransformed plant stained in parallel to those shown in a and b. (a–c, bar = 0.6 mm.) (d) Single flower cut transversely and photographed from above the cut surface, showing that staining is restricted to the anthers even when the other organs are cut to allow better substrate access. (e) Pollen washed out of the anthers of a transgenic plant before staining, and apparently segregating for GUS activity. (Bar = 100 μm.) (f and g) Seedlings of lines 47–9; 2.1 and 47–9; 2.3, respectively, at various stages of development. (h) Enlargments of the two youngest seedlings shown in g. (i and j) Dark-grown and light-grown seedlings of line 47–9; 2.3, respectively, photographed at the same magnification to allow direct comparison of root, hypocotyl, and cotyledon growth and staining.
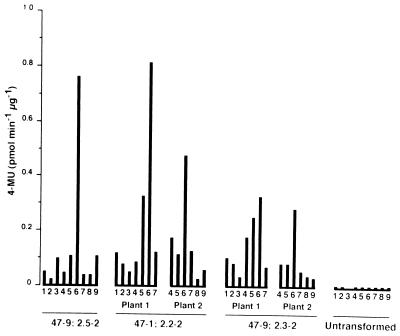
Figure 4.
GUS activity extracted from various organs, of three independent transgenic lines containing the At-RAB2–GUS fusion, 47–9; 2.5, 47–1; 2.2, and 47–9; 2.3, which stain with relatively high, medium, and low intensity, respectively, and from untransformed plants. The organs are: 1, roots; 2, leaves; 3, stems; 4, sepals; 5, petals; 6, stamens; 7, gynoecium; 8, siliques (ovules removed); 9, ovules. Activity is expressed as pmoles of 4-methyl-umbelliferone liberated per minute per microgram of total protein.
Wounding tissues prior to incubation did not increase the staining (i.e., Fig. 3d), suggesting that substrate access was not a significant factor. In contrast, plants expressing a CaMV-35S–GUS fusion showed staining of all organs, although some, notably the stem and gynoecium, had to be cut, and even then only cells near the wound were stained (not shown). We never observed staining in any tissue taken from untransformed plants (Table 1; Fig. 3c), from kanamycin-sensitive segregants of the At-RAB2–GUS transgenic lines, or from controls transformed by plasmids in which GUS was fused to an unrelated fragment of Arabidopsis genomic DNA.
The At-RAB2–GUS Fusion Is Activated Late in Pollen Development.
To establish the point during pollen development at which GUS activity appeared, GUS activity and pollen development were analyzed in each bud of an entire inflorescence, which represents a developmental series from apex to base. Inflorescences from three independent transgenic lines were studied. The first signs of GUS activity appeared in anthers around the onset of microspore mitosis, in one case in binucleate pollen grains after microspore mitosis, and in two others at the late microspore stage just prior to mitosis. In all three series, activity apparently increased with developmental age, and staining became most intense in the trinucleate stage. This is typical of class II pollen-expressed genes whose transcripts appear around the time of microspore mitosis and persist until maturity (22, 23). In contrast, class I transcripts are abundant during microspore development but decline after microspore mitosis. The results from one transgenic line are shown in Fig. 5.
Figure 5.
An inflorescence of a plant from line 47–9; 2.3 was stained for GUS activity for a prolonged period (20 hr) to enable the earliest signs of activity to be visualized. The younger buds were artificially opened to expose the interior, and each bud was photographed. (a–i; bars = 0.5 mm.) Buds were then incubated with the fluorescent DNA stain DAPI, a single anther was excised, and squash preparations from buds a–f were photographed under UV illumination, which revealed the gametophytic nuclei as bright blue fluorescent spots (b′–f′ below the corresponding bud; bars = 30 μm). (a) Apical buds left on the inflorescence axis and not examined individually. (b) Early mononucleate microspores; unstained. (c) Late mononucleate microspores; unstained. (d) Late mononucleate microspores; first signs of GUS activity. (e) Microspore mitosis has occurred (pollen grains show a condensed, brightly fluorescent generative nucleus as well as a larger, more diffuse vegetative nucleus); staining more pronounced. (f) Pollen mitosis has occurred (trinucleate pollen grains with two condensed generative nuclei that are readily visible, and a diffuse vegetative nucleus that is often hard to discern). (g–l) Further development of the flower. Staining reaches its maximum intensity in bud i. (j–l) Staining of the anther appears to wane as they shed pollen; stained pollen appears on the otherwise unstained stigma, and the vascular strand of the filament becomes stained.
As a test of the results obtained from the promoter–GUS analysis, the accumulation of At-RAB2 transcripts in flowers was analyzed. Pollen mitosis occurs in Arabidopsis flower buds of about 1 mm (24). Fig. 2B shows that At-RAB2 transcripts were more abundant in flower buds estimated to be greater than 1 mm than in those estimated to be less than 1 mm (compare lanes 3 and 4). It was reasoned that if this increase in At-RAB2 transcript abundance in older flowers is attributable to At-RAB2 expression in maturing pollen grains, a similar increase would not be seen in the buds of the Arabidopsis ms1 mutant whose microspores decay soon after release from tetrads (25). This was indeed the case (Fig. 2B, compare lanes 1 and 2 with lanes 3 and 4), supporting the conclusion from the GUS expression data that At-RAB2 is expressed predominantly in post-mitotic pollen grains.
Expression of the At-RAB2–GUS Fusions in Seedlings.
Other than pollen, the only tissues to show reproducible staining for GUS activity were those in the growing organs of newly emerged seedlings. At emergence, staining was observed in all organs but was usually most intense in the cotyledons (Fig. 3 f–h). Staining persisted while the cotyledons were growing but was lost as they approached full expansion. Cotyledons of seedlings containg a CaMV-35S–GUS fusion were efficiently stained even at full expansion, indicating that substrate access does not become limiting in these tissues. Seedlings at this stage from several At-RAB2–GUS lines (four of seven tested) showed enhanced staining of the crown and root (Fig. 3f), though the root tips were usually not stained. In some cases enhanced staining at the base of the hypocotyls was also observed (Fig. 3f). Staining persisted longest in the crown and the root but was eventually lost from these tissues too. These data suggested a transient increase in At-RAB2 transcription during the early stages of seedling growth. Fig. 2C shows that At-RAB2 transcripts were present in the unimbibed seeds, and that maximal transcript abundance was observed in whole seedlings 4 days after imbibition at the point when cotyledons were first becoming visible; cotyledons had become fully expanded by 8 days.
As mentioned above, staining for GUS activity was sometimes observed at the base of hypocotyls, and this seemed particularly evident in seedlings that showed the greatest hypocotyl growth. This observation was reinforced by staining seedlings that had germinated in darkness. Growth under these conditions primarily involves hypocotyl extension with relatively little root growth and minimal expansion of the cotyledons. Immediately after germination GUS staining was similar in dark-grown and light-grown seedlings, but after further growth in darkness it was most prominent and persistent in the elongating hypocotyls (Fig. 3i). This contrasts with light grown seedlings (compare Fig. 3 i and j), where hypocotyl growth and staining are lost early, and suggests that in these cells the At-RAB2 promoter can respond to physiological rather than purely tissue-specific signals. After further growth in darkness, staining intensity was much reduced in all tissues.
DISCUSSION
The transport of membrane vesicles between various compartments of the endomembrane system is central to the growth, differentiation, and functioning of plant cells. An understanding of the processes involved in vesicle transport is required to understand the biosynthesis of such important structures as the cell wall, plasma membrane, vacuole, and tonoplast.
Many intracellular vesicle transport pathways in mammalian and yeast cells are dependent upon one or more GTPases of the YPT/Rab family (2, 5). This family is part of the superfamily of small GTPases that are structurally related to p21-ras. (1). Much of the Rab family appears to be conserved in sequence and function among eukaryotes (2, 5). A number of reports have established that several members of this gene family are present in higher plants (6, 8, 9, 26), though some subclasses, including Rab2, have not been reported. It seems that no Rab2 homolog is present in the Saccharomyces cerevisiae genome (12), yet in mammals Rab2 appears to be necessary for membrane traffic between ER and Golgi (11). We have identified an Arabidopsis locus that encodes a protein with high sequence similarity to mammalian Rab2. Saalbach and Thielmann (27) have cited unpublished evidence that Vicia faba (field bean) also contains sequences homologous to Rab2.
The amino acid sequences of the Arabidopsis and animal Rab2 homologs share the effector region that typically is conserved only within specific subclasses of the YPT/Rab family (5, 28). They also share a second subclass-specific motif (29) in a region that appears to carry a second major determinant of Rab subclass specificity (30–32). It has been suggested (33) that the Volvox carterii yptV4 locus may be involved in endocytosis based on a comparison of part of its effector region with Rab4. However, we suggest that yptV4 is probably related to higher plant and animal Rab2 based on overall sequence similarity and the presence of the Rab2 specificity domains.
The predicted C-terminal residues CCG fit the consensus for plant YPT/Rab proteins (6–8). One of the corresponding cysteine residues at the C termini of YPT1p and SEC4p in S. cerevisiae is modified by the addition of a C20 prenyl group that facilitates membrane anchoring and biological activity (34, 35) and similar modifications have been reported for one plant Rab subclass (36).
At-RAB2 promoter fusions to the GUS reporter gene revealed strong histochemical staining only in pollen and young seedlings. The promoter of At-RAB2 contains the motifs 5′-TGTGG-3′ and 5′-TTGTGA-3′ situated 189 bp and 172 bp, respectively, upstream of the putative TATA box; these motifs are required for the expression of three tomato genes in post-mitotic pollen (37) and are found within 500 bp of the transcription start in maize and Petunia class II pollen genes (37–40). Adjacent to the 5′-TTGTGA-3′ motif, the At-RAB2 promoter also contains the motif 5′-AAATGA-3′ implicated in pollen-specific expression of the tobacco gene NTP303 and its Brassica homolog Bp10. However, the precise role of these promoter elements in directing expression in pollen is unclear (41).
We do not consider the upstream sequences of At-RAB2 to be strictly anther and seedling “specific.” First, in those lines which showed the highest levels of expression, some staining of tissues in other organs was also observed. Yet, these lines conformed to the general pattern, with maximal staining in pollen and seedlings. Furthermore, reducing the incubation time or including an inhibitor of GUS activity [K3Fe(CN)6; ref. 15] produced staining patterns similar to those of the other lines (data not shown), suggesting that these lines differed from the others principally in having higher levels of expression rather than altered or deregulated patterns of expression. Second, measurements of extractable GUS activity showed that although anthers contained the most activity all the other organs possessed activity considerably above that of controls even in plants where histochemical staining was apparent only in pollen and seedlings (e.g., 47–9; 2.3). Finally, At-RAB2 transcripts were detected in RNA from all organs tested.
The expression pattern of At-RAB2 suggests that it may be involved in a process that is required to some extent by most cells but is of particular significance in mature pollen and young seedlings. Mammalian Rab2 is located in a compartment between the ER and Golgi (10) and functions in transport between these two organelles (11). It may be that At-Rab2 performs a similar function. Indeed, there is a notable correspondence between the expression pattern of At-RAB2–GUS and the apparent requirements of various plant cell types for ER-to-Golgi traffic. These requirements appear to differ considerably between different plant cell types (42–45), though the evidence for this is largely circumstantial and derived from studies of the biosynthetic activities and ultrastructure of the ER and Golgi in plants. It appears that the majority of cells in the adult plant have only a minimal requirement for ER–Golgi traffic. In most such cells, putative ER–Golgi transport intermediates have yet to be identified, secretory rates are low, and the principal secreted material is complex cell wall polysaccharide, which is thought to be synthesized entirely in the Golgi with no known precursor from the ER (43, 45, 46). Membrane flow to the cell surface arising from transport of secretory vesicles is postulated to be balanced by recycling of membrane to the Golgi, obviating the need for stoichiometric membrane supply from the ER (43, 44). Thus, ER–Golgi traffic in adult tissues may function primarily in endomembrane maintenance, which may explain the widespread but comparatively weak expression of At-RAB2.
The ER-to-Golgi pathway is thought to be particularly important to cells that must undergo rapid growth and/or membrane biogenesis (44, 45, 47). The tissues most notable in this respect are germinating seedlings and pollen grains (44, 48, 49), both of which show pronounced At-RAB2 expression. In seedlings, the different expression patterns of At-RAB2–GUS in light and darkness suggest that the signals that influence its expression can be physiological rather than purely positional. Previously, Nagano et al. (50) have shown that pra2, a distant Rab11 homolog in pea, localizes to the elongation zone of etiolated epicotyls. As pollen grains mature they accumulate an extensive complement of ER and Golgi membranes that will be required to sustain pollen tube growth after germination (49). We suggest that At-RAB2 is transcribed in maturing pollen in preparation for germination. Indeed, other class II pollen genes are also thought to be required for pollen tube growth rather than for development of the grain (23, 24).
Peripheral mucilage-secreting cells of root tips also show high secretory vesicle production rates (42, 44, 46). Yet in Arabidopsis transgenic for At-RAB2–GUS they were not preferentially stained. However, this is consistent with the proposed function of At-Rab2, as these cells are not expected to have unusually high rates of ER–Golgi traffic because the mucilage they secrete is primarily Golgi-derived polysaccharide. If At-Rab2 does function in ER–Golgi traffic, it is perhaps anomalous that we found little evidence of enhanced expression in and around either the root or shoot meristems. It may be that the background expression level suffices; however, there is evidence from analysis of expressed sequence tags (51) and from our own observations for the existence of other Rab2 homologs in Arabidopsis, and their expression is under investigation.
Acknowledgments
We wish to thank Ulrike Klosse and Stephanie Rechmann for the automated sequence analysis, Riki Putnoky for care of the plants, John Baker for photographs, Drs. P. Bariola and I. Graham for the eIF4A plasmid, and various colleagues for comments. This work was funded in part by a Royal Society Research Grant to I.M. and by the Deutsche Forschungsgemeinschaft to K.P. (279/6-1). V.Z. was supported by the E.C. Go West program and GACR Grant 204 195/12G.
Note Added in Proof.
While this article was under review, the sequence and geranylation of a second Arabidopsis Rab2 homolog was reported (52) and a Brassica Rab2 homolog was reported (53).
Footnotes
References
- 1.Valencia A, Chardin P, Wittinghoffer A, Sander C. Biochemistry. 1991;30:4639–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 2.Pfeffer S. Curr Opin Cell Biol. 1994;6:522–526. doi: 10.1016/0955-0674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 3.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 4.Sollner T, Whitehart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 5.Zerial M, Stenmark H. Curr Opin Cell Biol. 1993;5:613–620. doi: 10.1016/0955-0674(93)90130-i. [DOI] [PubMed] [Google Scholar]
- 6.Terryn N, Van Montagu M, Inzé D. Plant Mol Biol. 1993;22:143–152. doi: 10.1007/BF00039002. [DOI] [PubMed] [Google Scholar]
- 7.Palme K, Diefenthal T, Vingron M, Sander C, Schell J. Proc Natl Acad Sci USA. 1992;89:787–791. doi: 10.1073/pnas.89.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagano Y, Murai N, Matsuno R, Sasaki Y. Plant Cell Physiol. 1993;34(3):447–455. [PubMed] [Google Scholar]
- 9.Bednarek S Y, Reynolds T L, Schroeder M, Grabowski R, Hengst L, Gallwitz D, Raikhel N V. Plant Physiol. 1994;104:591–596. doi: 10.1104/pp.104.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavrier P, Parton R G, Hauri H P, Simons K, Zerial M. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- 11.Tisdale E J, Bourne J R, Khosravi-Far R, Der C J, Balch W E. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh S, Barrel B. Trends Genet. 1996;12:276–277. doi: 10.1016/0168-9525(96)60029-8. [DOI] [PubMed] [Google Scholar]
- 13.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Green/Wiley Interscience; 1989. [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Jefferson R A. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 16.Koncz C, Schell J S. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 17.Valvekens D, Van Montagu M, Van Lijsebettens M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemper E, Grevelding C, Schell J, Masterson R. Plant Cell Rep. 1992;11:118–121. doi: 10.1007/BF00232162. [DOI] [PubMed] [Google Scholar]
- 19.Agterberg M, Vandie I, Yang H, Andriessen J A, Vantetering A, Vandeneijnden D H, Ploegh H L. Eur J Biochem. 1993;217:241–246. doi: 10.1111/j.1432-1033.1993.tb18239.x. [DOI] [PubMed] [Google Scholar]
- 20.Brown J. Nucleic Acids Res. 1986;14:9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascarenhas J P. Annu Rev Plant Physiol. 1990;41:317–338. [Google Scholar]
- 23.Scott R, Hodge R, Paul W, Draper J. Plant Sci. 1991;80:167–191. [Google Scholar]
- 24.Eady C, Lindsey K, Twell D. Plant J. 1994;5:543–550. [Google Scholar]
- 25.van der Veen J H, Wirtz P. Euphytica. 1968;17:371–377. [Google Scholar]
- 26.Terryn N, Arias M B, Engler G, Tiré C, Villaroel R, Van Montagu M, Inzé D. Plant Cell. 1993b;5:1761–1769. doi: 10.1105/tpc.5.12.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saalbach G, Thielmann J. J Plant Physiol. 1995;145:665–673. [Google Scholar]
- 28.Haubruck H, Engelke U, Mertins P, Gallwitz D. EMBO J. 1990;9:1957–1962. doi: 10.1002/j.1460-2075.1990.tb08323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore I R, Schell J S, Palme K. Trends Biochem Sci. 1995;20:10–12. doi: 10.1016/s0968-0004(00)88939-2. [DOI] [PubMed] [Google Scholar]
- 30.Brennwald P, Novick P. Nature (London) 1993;362:560–563. doi: 10.1038/362560a0. [DOI] [PubMed] [Google Scholar]
- 31.Dunn B, Stearns T, Botstein D. Nature (London) 1993;362:563–565. doi: 10.1038/362563a0. [DOI] [PubMed] [Google Scholar]
- 32.Stenmark H, Valencia A, Martinez O, Ullrich O, Goud B, Zerial M. EMBO J. 1994;13:575–583. doi: 10.1002/j.1460-2075.1994.tb06295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabry S, Jacobsen A, Huber H, Palme K, Schmitt R. Curr Genet. 1993;24:229–240. doi: 10.1007/BF00351797. [DOI] [PubMed] [Google Scholar]
- 34.Molenaar C, Prange R, Gallwitz D. EMBO J. 1988;7:971–976. doi: 10.1002/j.1460-2075.1988.tb02903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinsella T, Maltese W. J Biol Chem. 1992;267:3940–3945. [PubMed] [Google Scholar]
- 36.Loraine A E, Yalovsky S, Fabry S, Gruissem W. Plant Physiol. 1996;110:1337–1347. doi: 10.1104/pp.110.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Twell D, Yamaguchi J, Wing R A, Ushiba J, McCormick S. Genes Dev. 1991;5:496–507. doi: 10.1101/gad.5.3.496. [DOI] [PubMed] [Google Scholar]
- 38.van Tunen A J, Hartman S A, Mur L A, Mol J N M. Plant Mol Biol. 1989;12:539–551. doi: 10.1007/BF00036968. [DOI] [PubMed] [Google Scholar]
- 39.Hamilton D A, Bashe D M, Stinson J R, Mascarenhas J P. Sex Plant Reprod. 1989;2:208–212. [Google Scholar]
- 40.Dubald M, Barakate A, Mandaron P, Mache R. Plant J. 1993;4:781–791. doi: 10.1046/j.1365-313x.1993.04050781.x. [DOI] [PubMed] [Google Scholar]
- 41.Wetterings K, Schrauwen J, Wullems G, Twell D. Plant J. 1995;8:55–63. doi: 10.1046/j.1365-313x.1995.08010055.x. [DOI] [PubMed] [Google Scholar]
- 42.Battey N H, Blackbourn H D. New Phytol. 1993;125:307–338. doi: 10.1111/j.1469-8137.1993.tb03883.x. [DOI] [PubMed] [Google Scholar]
- 43.Hawes C, Faye L, Satiat-Jeunemaitre B. In: Membranes: Specialised Functions in Plants. Smallwood M, Knox J P, Bowles D, editors. Oxford: BIOS; 1995. [Google Scholar]
- 44.Steer M W, O’Driscoll D. In: Endocytosis, Exocytosis and Vesicle Traffic in Plants. Hawes C R, Coleman J O D, Evans D E, editors. Cambridge, U.K.: Cambridge Univ. Press; 1991. pp. 129–142. [Google Scholar]
- 45.Moore P J, Staehelin L A. Planta. 1988;174:433–445. doi: 10.1007/BF00634471. [DOI] [PubMed] [Google Scholar]
- 46.Driouich A, Faye L, Staehelin L A. Trends Biochem Sci. 1993;18:210–214. doi: 10.1016/0968-0004(93)90191-o. [DOI] [PubMed] [Google Scholar]
- 47.Bolwell G P. Phytochemistry. 1988;27(5):1235–1253. [Google Scholar]
- 48.Phillips G D, Preshaw C, Steer M W. Protoplasma. 1988;145:59–65. [Google Scholar]
- 49.Steer M W, Steer J M. New Phytol. 1989;111:323–358. doi: 10.1111/j.1469-8137.1989.tb00697.x. [DOI] [PubMed] [Google Scholar]
- 50.Nagano Y, Okada Y, Narita H, Asaka Y, Sasaki Y. Proc Natl Acad Sci USA. 1995;92:6314–6318. doi: 10.1073/pnas.92.14.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooke R, et al. Plant J. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- 52.Bierman B, Randell S K, Crowell D N. Plant Mol Biol. 1996;31:1021–1028. doi: 10.1007/BF00040720. [DOI] [PubMed] [Google Scholar]
- 53.Kim W Y, Cheong N E, Lee D C, Lee K O, Je D Y, et al. Plant Mol Biol. 1996;31:783–792. doi: 10.1007/BF00019466. [DOI] [PubMed] [Google Scholar]