Abstract
Previous molecular analyses of mammalian evolutionary relationships involving a wide range of placental mammalian taxa have been restricted in size from one to two dozen gene loci and have not decisively resolved the basal branching order within Placentalia. Here, on extracting from thousands of gene loci both their coding nucleotide sequences and translated amino acid sequences, we attempt to resolve key uncertainties about the ancient branching pattern of crown placental mammals. Focusing on ≈1,700 conserved gene loci, those that have the more slowly evolving coding sequences, and using maximum-likelihood, Bayesian inference, maximum parsimony, and neighbor-joining (NJ) phylogenetic tree reconstruction methods, we find from almost all results that a clade (the southern Atlantogenata) composed of Afrotheria and Xenarthra is the sister group of all other (the northern Boreoeutheria) crown placental mammals, among boreoeutherians Rodentia groups with Lagomorpha, and the resultant Glires is close to Primates. Only the NJ tree for nucleotide sequences separates Rodentia (murids) first and then Lagomorpha (rabbit) from the other placental mammals. However, this nucleotide NJ tree still depicts Atlantogenata and Boreoeutheria but minus Rodentia and Lagomorpha. Moreover, the NJ tree for amino acid sequences does depict the basal separation to be between Atlantogenata and a Boreoeutheria that includes Rodentia and Lagomorpha. Crown placental mammalian diversification appears to be largely the result of ancient plate tectonic events that allowed time for convergent phenotypes to evolve in the descendant clades.
Keywords: Atlantogenata, Eutheria, Notolegia, phylogeny, vicariance
Phylogenetic analyses can elucidate the history of diversification within a group of organisms such as placental mammals (i.e., Placentalia) (1, 2). However, if the analyses use too few characters or taxa, an inaccurate phylogenetic tree can be obtained because of sampling error (3). Here we seek to reduce such error by using abundant character information from mammalian and other vertebrate genomes that have been completely or nearly completely sequenced. On examining phylogenetically the coding nucleotide sequences and translated amino acid sequences for thousands of genes, we attempt to resolve key uncertainties about the ancient branching order of crown placental mammals. Our results are promising, but uncertainties remain concerning the basal diversification of Placentalia. The practice of phylogenomics is still in its infancy and has yet to produce an authoritative model that infallibly predicts all of the real patterns of nucleotidesubstitutions within evolved genomes.
Among the placental mammals, phylogenetic branching events have been inferred by using either nucleotide sequence data (4–11) or rare insertion/deletion patterns (12–15). Many of the results recognize four primary eutherian groups: Afrotheria, Xenarthra, Laurasiatheria, and Euarchontoglires. Afrotherians (e.g., elephants, hyraxes, manatees, aardvarks, tenrecs, and allies) are a clade of mammals that originated in Africa, and whose extant members still mostly remain on that continent with the exception of Asian elephants and sirenians such as the Florida manatee. The Xenarthra includes the sloths, armadillos, and anteaters that today are restricted to South and Central America (although some Xenarthra, such as the nine-banded armadillo, have recently dispersed to North America). The Laurasiatheria (e.g., bats, eulipotyphlans, pangolins, carnivores, perrisodactyls, and cetartiodactyls) is a diverse clade including extant lineages that originated in the ancient northern continent of Laurasia. The Euarchontoglires includes the species from five living mammalian orders (e.g., primates, treeshrews, flying lemurs, rabbits, and rodents). This last group remains the most controversial, and a number of recent studies have suggested it is not valid (4, 6, 7).
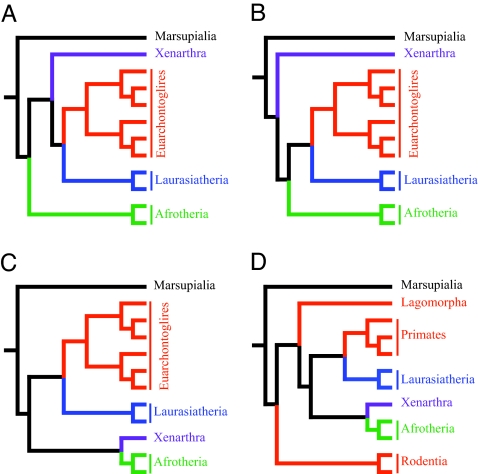
The studies that have identified and supported these primary groupings have been able to resolve the branching pattern within some of the clades, but a statistically robust determination of the branching order at the base of the placental clade continues to be elusive. Three primary hypotheses (Fig. 1 A–C) supporting the branching orders among the four major placental groups described above have been proposed. In the first scenario (9, 10, 16), Afrotherians split from the other three clades (Notolegia = Exafroplacentalia) at the base of the placental tree. In the second hypothesis (13), the Xenarthrans split from the other three clades (Epitheria). The third hypothesis (5, 11, 14, 17) proposes that Xenarthra and Afrotheria group together (Atlantogenata) to the exclusion of Laurasiatheria and Euarchontoglires (Boreoeutheria).
Fig. 1.
Alternative hypotheses regarding the branching order among placental mammals. (A) Afrotheria as sister taxon to the other placental mammalian clades. (B) Xenarthra as sister taxon to the other placental mammalian clades. (C) Xenarthra and Afrotheria group together to the exclusion of the other clades. (D) Murid rodents as sister taxon to the other placental mammalian clades; Glires is disrupted by the joining of Lagomorpha to the remaining placental clades.
Some studies have suggested a fourth hypothesis, that one of the groups, Euarchontoglires, is polyphyletic, and that Glires (rodents and lagomorphs) is also not monophyletic (Fig. 1D). Based on a parsimony analysis of nuclear-encoded genes, it was reported (8) that murid rodents as represented by mouse and rat were the sister group to all other placental mammals, and that rabbits were the next branching clade. More recently, a number of large-scale genomic studies (4, 6) have reported that primates grouped more closely to laurasiatherians than to Glires.
To test the four alternative hypotheses (Fig. 1), we constructed phylogenetic trees and performed topology tests based on a data set that included nucleotide and amino acid sequence data from the complete genome sequences of 11 mammalian species representing the major placental clades depicted in Fig. 1, a marsupial opossum, and two nonmammalian outgroups. The ingroup taxa include three primates (human, chimpanzee, and rhesus macaque), two rodents (mouse and rat), rabbit, dog, cow, armadillo, African elephant, tenrec, and the South American Gray Short-tailed opossum. Chicken and frog were used as outgroups to root the tree. To avoid common pitfalls associated with phylogenomic studies, we focused our analyses on conserved protein coding genes to reduce saturation and long-branch attraction effects. Our alignment method also preserved the codon reading frame for all loci and all taxa, so we were able to remove potentially saturated third codon positions from the parsimony analyses. Because all tree reconstruction methods fail to recover the “true” tree given certain conditions, we used four different inferential tree reconstruction methods: maximum parsimony (MP), maximum likelihood (ML), Bayesian, and neighbor-joining (NJ). The probabilistic methods (ML and Bayesian) may be more robust than the others to model assumption violation, although NJ can also often recover the true tree when the wrong model of sequence evolution is used. MP minimizes the amount of evolution (i.e., nucleotide substitutions) and in some cases, outperforms the probabilistic methods (18, 19). Although we would be encouraged if several of these tree reconstruction methods converged on just one of the four hypotheses, we realize that all methods might converge on an incorrect branching pattern for the basal diversification of Placentalia.
Results
When all human RefSeq mRNA transcripts (n = 25,556) were aligned to their putative orthologs, the result was a gapped alignment of 36 Mb. However, orthologs for most genes could not be found for all 14 taxa. Therefore, the analyses included only those loci for which our method could assign orthologous sequences for all 14 taxa. This reduced data set consisted of a multiple sequence alignment of 1,698 protein-coding loci with an alignment length of 1,443,825 bp, including insertions and deletions. The mean composition of nucleotide bases in this alignment was as follows: T = 22.8%, C = 23.8%, A = 27.8%, and G = 25.6%. A table showing the nucleotide base composition at each codon position for each taxon as well as the number of nucleotides analyzed is included in supporting information (SI) Table 3. Notably, for each base at each of the three codon positions, there are at most only very small compositional differences among the 14 taxa. A summary spreadsheet of the putative orthologs used in the main analyses (SI Table 4), as well as the accompanying alignment files are available as SI.
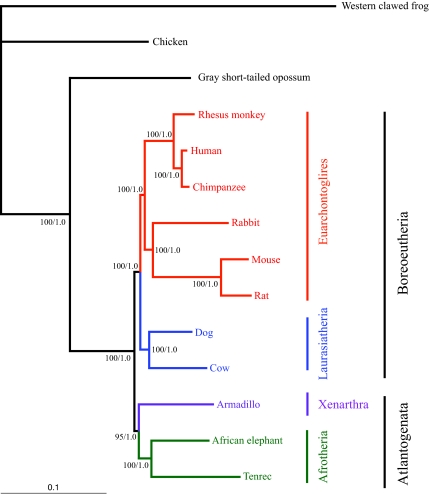
Fig. 2 depicts the optimal phylogenetic branching pattern among the taxa whether a Bayesian, ML, or MP phylogenetic tree reconstruction method is used with the coding nucleotide sequence data or the parsimony amino acid data. The branch lengths in Fig. 2 were obtained by ML analysis. Fig. 3 depicts the optimal NJ trees. The MP nucleotide tree has a length of 267,158 steps, and the ML scores for the tree topology are -lnL 7,238,023.807 (PAUP*) and -lnL 7,240,210.130 (PhyML). The branch support values for each of the clades are 100% (MP and PhyML bootstrap percent) or 1.0 (Bayesian posterior probability) at all nodes in the tree with the single exception of the Atlantogenata clade, which had a parsimony bootstrap value of 95% in the nucleotide analysis. The relatively short branch lengths seen in the catarrhine and, more specifically, the ape clade also provide further evidence for the hominid slowdown hypothesis (20, 21).
Fig. 2.
Phylogenetic relationships among major placental groups. Optimal tree topology obtained by MP using first and second codon positions (267,158 steps), PAUP* ML (-ln L = 7,238,023.807), PhyML ML (-ln L = 7,240,210.130) and Bayesian approaches, based on the 1,443,825-bp alignment. PhyML bootstrap supports equal 100% for all nodes. Bayesian posterior probabilities equal 1.0 for all nodes. MP bootstrap supports equal 100% for all nodes with the exception of a 95% value at one node. Branch lengths reflect the likelihood distances calculated by PAUP* Ver. 4.0b10 using the GTR + I + Γ model chosen by ModelTest.
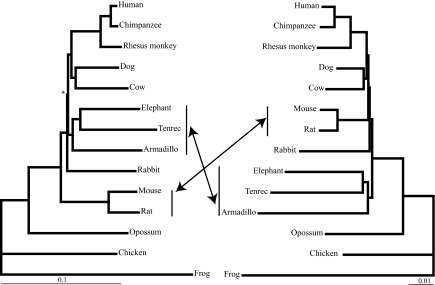
Fig. 3.
NJ analyses of amino acid and nucleotide data sets. Optimal tree topology and branch lengths obtained by NJ analyses of nucleotides (Left) using the maximum composite likelihood distance and amino acid (Right) using the JTT distance. Bootstrap values of all nodes were 100% for 1,000 replicates, except when indicated by * (=97%). An identical tree topology to that shown for amino acids was obtained when only first and second codon positions were used.
The NJ tree confirms the presence of the Atlantogenata clade with bootstrap support of 100%, although it differs from the other three methods by not supporting Euarchontoglires or Glires as monophyletic, indicating rodents as the first branching placental clade and depicting Laurasiatheria as the sister group to primates (Fig. 3). Parsimony and NJ results from the translated amino acid sequences both depict a basal split between Atlantogenata and Boreoeutheria. Whereas the parsimony amino acid and nucleotide sequence data results show an identical topology, the NJ amino acid tree depicts a boreoeutherian clade not detected by using nucleotide sequence data and indicates Lagomorpha is sister to a clade that includes the remaining Boreoeutheria (Fig. 3). Notably, with nucleotide sequences, when only first and second codon positions (positions less likely to be saturated by superimposed mutations) are retained, the NJ tree again depicts the basal Placentalia divergence to be between Atlantogenata and Boreoeutheria.
We conducted parsimony and likelihood topology tests on the phylogenetic trees constructed by using the coding nucleotide sequence data (Tables 1 and 2). The likelihood tests clearly indicate the tree depicted in Figs. 1C and 2 is the optimal tree (P < 0.0001; Table 1). The trees depicted in Fig. 1 A, B, and D are rejected as being suboptimal. Results of parsimony tests also reject the topologies depicted in Fig. 1 A and D in favor of Fig. 1C (Table 2); however, the topology in Fig. 1B cannot be rejected (P = 0.0897; Templeton test). The parsimony scores from these two topologies differ by only 73 steps.
Table 1.
Likelihood tests of alternative topologies
| Shimodaira–Hasegawa test | |||
|---|---|---|---|
| Tree | -ln L | Diff -ln L | P |
| A | 7,239,065.38 | 1,041.57228 | 0.000* |
| B | 7,239,235.261 | 1,211.45404 | 0.000* |
| C | 7,238,023.807 | Best | |
| D | 7,243,526.067 | 5,502.25965 | 0.000* |
Shimodaira–Hasegawa test using RELL bootstrap (one-tailed test).
Number of bootstrap replicates, 1,000.
* P < 0.05.
Table 2.
Parsimony tests of alternative topologies
| Tree | Length | Kishino–Hasegawa test |
Templeton test |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Difference | SD (diff) | t | P* | Rank sums† | N | z | P‡ | ||
| A | 267,548 | 390 | 39.16432 | 9.958 | <0.0001* | 738,335 | 1,534 | −9.9575 | <0.0001* |
| −439,010 | |||||||||
| B | 267,231 | 73 | 43.02321 | 1.6968 | 0.0897 | 890,812 | 1,851 | −1.6968 | 0.0897 |
| −823,214 | |||||||||
| C | 267,158 | Best | Best | ||||||
| D | 268,181 | 1,023 | 71.25951 | 14.356 | <0.0001* | 5,834,116.5 | 4,365 | −14.4658 | <0.0001* |
| −3,694,678.5 | |||||||||
*Probability of getting a more extreme T value under the null hypothesis of no difference between the two trees (two-tailed test). Asterisked values in table indicate significant difference at P < 0.05.
†Wilcoxon signed-ranks test statistic is the smaller of the absolute values of the two rank sums.
‡Approximate probability of getting a more extreme test statistic under the null hypothesis of no difference between the two trees (two-tailed test). Asterisked values in table indicate significant difference at P < 0.05.
Discussion
Our data set of 1,698 protein-encoding loci contained putative orthologous sequences from every one of the genomes of the 12 chosen mammal species plus the chicken and frog outgroups. The detection of orthologs among such a wide range of vertebrate taxa suggests this data set represents relatively slowly evolving DNA capable of revealing the ancient branching order of Placentalia. We found that Afrotheria and Xenarthra form a sister group (i.e., Atlantogenata) to a clade comprising Euarchontoglires and Laurasiatheria (i.e., Boreoeutheria) when MP, ML, or Bayesian approaches are used. The NJ approach also depicts an Atlantogenatan clade whether amino acid or nucleotide sequences are used to infer the tree; however, the NJ tree based on nucleotide sequences is alone in depicting a basal separation of Rodentia (murids) from all other placental mammals. The differences between this nucleotide NJ tree topology compared with MP/ML/Bayesian tree topology and also the amino acid NJ tree topology suggests methodological failure of at least one of these methods. In terms of either parsimony or likelihood criteria, the topology tests we conducted strongly reject the nucleotide NJ tree. The high bootstrap support for the MP/ML/Bayesian (Fig. 2) but also for the NJ nucleotide tree (Fig. 3) further indicates that one or the other tree-reconstruction approaches is inappropriate for the data and produces an incorrect tree because of systematic error (22–26).
The main biases that can cause systematic error in tree reconstruction are nucleotide compositional bias, long-branch attraction, and heterotachy (22, 24). Compositional bias has been shown to affect phylogeny reconstruction such that subsets of unrelated species that have converged on similar nucleotide compositions are grouped together erroneously. In a phylogenomic study of yeast orthologs (27), compositional bias was shown to lead to inconsistency in distance methods but not ML. In the present data set, compositional bias does not appear to be a problem, because the nucleotide compositions are similar among the taxa sampled. Especially noteworthy is this compositional similarity is manifested at each of the three codon positions (SI Table 3). Nonetheless, we removed third codon positions from parsimony analysis, because third positions are more likely the source of homoplastic substitutions that can cause long-branch attraction.
Long-branch attraction is a classic phylogenetic problem that incorrectly unites long branches together in a clade (28). It is a particular problem in MP, which fails to correct for parallel changes on long branches (25, 26). Long-branch attraction can cause systematic error in all methods used in this study, but there are data that suggest adding more taxa can break up long branches, reducing the probability of error (29). Interestingly, an NJ analysis of the Murphy et al. (9) data set composed of 44 mammalian taxa detected, as in the original study, an Afrotherian clade as sister to all other placental mammals (SI Text), rather than the rodent first separation detected by the nucleotide NJ analysis (all codon positions) performed in this study. The tree topology also shows a monophyletic Glires and Euarchontoglires. To so analyze the Murphy data set by NJ demonstrates long-branch attraction effect gets drastically reduced by denser taxon sampling (which breaks up the rodent long branch). Indeed, if the NJ analysis of the Murphy data set includes only the taxa represented in this study, a “rodent first” topology is once again recovered. The present study sampled more taxa than other recent mammalian phylogenomic studies (4, 6) and is therefore less likely to be affected by the long-branch attraction problem. This point is borne out by the finding that our data set recovered identical topologies to those reported by Huttley et al. (6) and Cannarrozzi et al. (4) when we limited our data to include only the taxa in those studies (see SI Text). Furthermore, the parsimony nucleotide results were obtained by using only first and second codon positions, which are less subject to the parallel changes that contribute to the long-branch attraction problem, and the topology obtained was identical to that recovered when MP was applied to the translated amino acid sequences (which also reduces the likelihood of erroneously identifying homoplasies as synapomorphies).
Variation in substitution rate at a single base or amino acid position over evolutionary time is referred to as heterotachy and can result in phylogenetic artifacts (30, 31). Errors resulting from heterotachy are difficult to detect with the methods we used; however, it has been shown that MP methods are sometimes less sensitive to heterotachy than are the probabilistic ML and Bayesian techniques (19), and in this study, results were congruent among these three methods. Nevertheless, further investigation of the cladistic relationships among mammals is now possible, because more eutherian genomes are available (e.g., platypus, cat, horse, bat, galago, treeshrew, and guinea pig). The availability of these genomes will allow further testing of the Atlantogenata/Boreoeutheria split and, within Boreoeutheria, the Glires hypothesis depicted in the majority of analyses in this study.
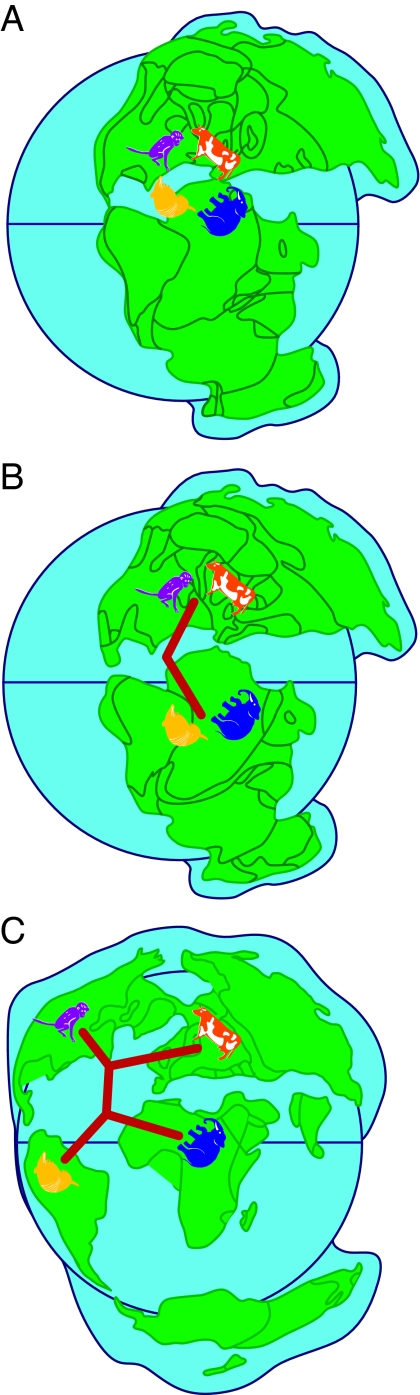
In our view, the weight of evidence now points to a sister group relationship between Atlantogenata and Boreoeutheria, and a clear scenario of biogeographic diversification emerges (Fig. 4). In this scenario, the placental mammals would have been subdivided into two lineages when the spreading Tethyan seaway widely separated Gondwana in the south from Laurasia in the north during the Cretaceous (32, 33). This process divided the initial members of the clades Boreoeutheria in the north from their southern atlantogenatan counterparts. Also, later in the Cretaceous, the disconnection of the African and South American landmasses ≈100 million years ago would have resulted in vicariance within Atlantogenata. This vicariant separation resulted in the clades Afrotheria in Africa and Xenarthra in South America. The mode of diversification between Laurasiatheria and Euarchontoglires remains murky, and it is unclear whether this was primarily because of vicariance between North America and Eurasia, some other vicariant event, or dispersal. Some remarkably similar morphological features that have emerged among the mammalian clades in the different geographic areas led previous workers to group divergent taxa together in polyphyletic assemblages (e.g., ungulates) based on convergent evolution of hoofs or to assume that features emerged only once (e.g., the variant types of gross anatomy in the placenta). With the availability of the mammalian genome sequences ever accumulating, it is now possible to design and test phylogenetic hypotheses about the genetic underpinnings of these and other important aspects of mammalian phenotypes.
Fig. 4.
Plate tectonics explain the diversification of the major placental clades. The most parsimonious reconstruction of placental mammalian diversification, given the phylogenetic findings (i.e., Fig. 2), is depicted. The four major clades of placental mammals are Afrotheria (represented by elephant), Xenarthra (represented by armadillo), Laurasiatheria (represented by cow), and Euarchontoglires (represented by monkey). (A) Eutheria originated on the supercontinent of Pangaea during the Jurassic. At this time, the four placental clades had not diverged from each other. (B) The initial split between placental clades occurred during the Creataceous when Gondwana and the northern continent of Laurasia became widely divided. This separated the southern mammalian clade Atlantogenata (Afrotheria and Xenarthra) from the northern clade Boreoeutheria (Laurasiatheria and Euarchontoglires). (C) In the south, the late Cretaceous separation of Africa and South America is coincident with the divergence of Afrotheria and Xenarthra. The separation of Laurasiatheria and Euarchontoglires also occurred in the north, and diversification among placental mammalian orders was complete by the early Cenozoic. Maps are adapted from ref. 53.
Materials and Methods
Details of analysis parameters and settings are available in SI Text.
Data Composition.
Orthology extraction and multiple sequence alignment were performed by using Online Codon-Preserved Alignment Tool (OCPAT), an in-house-developed tool that combines the BLAST (34) and Clustal (35) algorithms and preserves the correct protein coding reading frame in all taxa aligned. The tool is available at http://homopan.wayne.edu/Pise/ocpat.html. Details of the analysis pipeline are provided at http://homopan.wayne.edu/ocpat/index.html (54). Taxa included in the study were Homo sapiens (human) (36), Pan troglodytes (chimpanzee) (37), Macaca mulatta (Rhesus monkey) (38), Mus musculus (mouse) (39), Rattus norvegicus (rat) (40), Oryctolagus cuniculus (rabbit), Canis familiaris (dog) (41), Bos taurus (cow), Dasypus novemcinctus (armadillo), Loxodonta africana (African elephant), Echinops telfairi (tenrec), Monodelphis domestica (gray short-tailed opossum) (42), Gallus gallus (chicken) (43), and Xenopus tropicalis (Western clawed frog). For the analysis, all data we used were updated at the end of August 2006. Ornithorhynchus anatinus (platypus) and other recently completed draft genomes were not included. Nucleotide composition was calculated by using MEGA 4.0 (44).
Phylogenetic Analyses.
ML analysis.
ML analyses were conducted in PAUP* Ver. 4.0b10 (45) by using Model settings determined by the program ModelTest Ver. 3.7 (46), as chosen by Akaike Information Criterion (AIC). These settings correspond to the general time reversible (GTR) + Γ + I model with four rate categories. Assumed nucleotide frequencies were: A = 0.27630, C = 0.24200, G = 0.25440, and T = 0.22730. The assumed proportion of invariable sites = 0.3082 and the distribution of rates at variable sites = gamma (discrete approximation) with a shape parameter (α) = 0.6954. Our heuristic analysis included 10 random sequence additions and tree-bisection-reconnection (TBR) tree searching. Complete settings are available in SI Text. Additional analyses were conducted in a new beta version (3.0) of PhyML (47), which includes fast subtree pruning and regrafting (SPR) tree search (48). We first ran PhyML with an SPR search from 10 random starting trees and with GTR + Γ + I model with four rate categories but without bootstrap. The proportion of invariant sites, shape parameter of the gamma distribution and GTR parameters were estimated from the data. Then, we performed a bootstrap analysis with 100 replicates by using BioNJ trees as starting points, SPR tree search, and the model parameter values estimated in the first run. All inferred trees (i.e., 10 with random starting trees and 100 with resampled data) were identical.
Bayesian analysis.
The parallel MrBayes [Ver. 3.1.2 (49–51)] analysis was carried out by using the resources of the Computational Biology Service Unit from Cornell University (Ithaca, NY).
With 14 taxa and a concatenated sequence of 1,443,825 bases, the parallel processes ran for ≈1 week for 1 million generations of Markov-Chain Monte Carlo. We chose to assume partition homogeneity rather than using mixed models, which potentially would suffer from overparameterization from the 1,600+ loci included in the analysis. For the 1.4-Mb data set, we also ran the analysis using BayesPhylogenies (52). In this case, we assumed a single GTR pattern.
MP analysis.
We conducted a heuristic search in PAUP* Ver. 4.0b10 consisting of 100 random addition sequence replicates using the tree-bisection-reconnection (TBR) branch-swapping algorithm. We excluded third codon positions in parsimony analysis because of potential saturation at those sites. Thus, 481,275 characters were excluded, leaving a data set that included the remaining 962,550 characters. In addition to searching for the optimal tree, we conducted a bootstrap analysis that was based on 1,000 pseudoreplicates with 10 random addition sequence replicates per pseudoreplicate. The TBR algorithm was also used in this analysis. We also conducted an MP analysis on the translated amino acid sequences. In addition to the full complement of taxa, we ran parsimony analyses to reflect the taxon sampling in refs. 4 and 6.
NJ analysis.
We conducted an NJ search in PAUP* Ver. 4.0b10 with 1,000 full heuristic bootstrap replicates using the ML distances as selected by ModelTest and in MEGA 4.0 using the ML composite distance. We also conducted an NJ bootstrap analysis (1,000 replicates) in MEGA on the translated amino acid sequences using the Jones–Taylor–Thornton distance. In addition to the full complement of taxa, we ran NJ analyses to reflect the taxon sampling in refs. 4 and 6.
Supplementary Material
Acknowledgments
Andrew Meade (University of Reading, Reading, U.K.) provided access to computing resources, and part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University, which is partially funded by Microsoft Corporation. Munirul Islam (Wayne State University, Detroit, MI) provided computational support. We thank Wen-Hsiung Li (University of Chicago, Chicago, IL), Wilfried W. de Jong (University of Nijmegen, Nijmegen, The Netherlands), Nathaniel M. Pearson (Wayne State University), two anonymous reviewers, and the PNAS editorial board for comments and suggestions that greatly improved the manuscript. This research was supported in part by the Intramural Research division of National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services and National Science Foundation Grant BCS-0550209. We acknowledge the sources of the unpublished genomic data, including: Baylor College of Medicine Human Genome Sequencing Center (cow; http://www.hgsc.bcm.tmc.edu/projects); the Broad Institute (rabbit, elephant, tenrec, and armadillo); and the U.S. Department of Energy Joint Genome Institute (frog).
Abbreviations
- MP
maximum parsimony
- ML
maximum likelihood
- NJ
neighbor-joining.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704342104/DC1.
References
- 1.McKenna MC, Bell SK, Simpson GG. Classification of Mammals Above the Species Level. New York: Columbia Univ Press; 1997. [Google Scholar]
- 2.Lecointre G, Le Guyader H. The Tree of Life: A Phylogenetic Classification. Cambridge, MA: Belknap Press of Harvard Univ Press; 2006. [Google Scholar]
- 3.Donoghue MJ, Cracraft J. In: Assembling the Tree of Life. Cracraft J, Donoghue MJ, editors. Oxford, UK: Oxford Univ Press; 2004. pp. 1–4. [Google Scholar]
- 4.Cannarozzi G, Schneider A, Gonnet G. PLoS Comput Biol. 2007;3:e2. doi: 10.1371/journal.pcbi.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douady CJ, Chatelier PI, Madsen O, de Jong WW, Catzeflis F, Springer MS, Stanhope MJ. Mol Phylogenet Evol. 2002;25:200–209. doi: 10.1016/s1055-7903(02)00232-4. [DOI] [PubMed] [Google Scholar]
- 6.Huttley GA, Wakefield MJ, Easteal S. Mol Biol Evol. 2007;24:1702–1730. doi: 10.1093/molbev/msm094. [DOI] [PubMed] [Google Scholar]
- 7.Kullberg M, Nilsson MA, Arnason U, Harley EH, Janke A. Mol Biol Evol. 2006;23:1493–1503. doi: 10.1093/molbev/msl027. [DOI] [PubMed] [Google Scholar]
- 8.Misawa K, Nei M. J Mol Evol. 2003;57(Suppl 1):S290–S296. doi: 10.1007/s00239-003-0039-7. [DOI] [PubMed] [Google Scholar]
- 9.Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, et al. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 10.Nikolaev S, Montoya-Burgos JI, Margulies EH, Program NC, Rougemont J, Nyffeler B, Antonarakis SE. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rheede T, Bastiaans T, Boone DN, Hedges SB, de Jong WW, Madsen O. Mol Biol Evol. 2006;23:587–597. doi: 10.1093/molbev/msj064. [DOI] [PubMed] [Google Scholar]
- 12.de Jong WW, van Dijk MAM, Poux C, Kappe G, van Rheede T, Madsen O. Mol Phylogenet Evol. 2003;28:328–340. doi: 10.1016/s1055-7903(03)00116-7. [DOI] [PubMed] [Google Scholar]
- 13.Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishihara H, Hasegawa M, Okada N. Proc Natl Acad Sci USA. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asher RJ. In: The Rise of Placental Mammals: Origins and Relationships of the Major Extant Clades. Rose KD, Archibald JD, editors. Baltimore, MD: Johns Hopkins Univ Press; 2005. pp. 50–70. [Google Scholar]
- 17.Waters PD, Dobigny G, Waddell PJ, Robinson TJ. PLoS ONE. 2007;2:e158. doi: 10.1371/journal.pone.0000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 19.Kolaczkowski B, Thornton JW. Nature. 2004;431:980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- 20.Bailey WJ, Fitch DH, Tagle DA, Czelusniak J, Slightom JL, Goodman M. Mol Biol Evol. 1991;8:155–184. doi: 10.1093/oxfordjournals.molbev.a040641. [DOI] [PubMed] [Google Scholar]
- 21.Elango N, Thomas JW, Yi SV. Proc Natl Acad Sci USA. 2006;103:1370–1375. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delsuc F, Brinkmann H, Philippe H. Nat Rev. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 24.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 26.Yang Z. Computational Molecular Evolution. Oxford: Oxford Univ Press; 2006. [Google Scholar]
- 27.Phillips MJ, Delsuc F, Penny D. Mol Biol Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Syst Zool. 1978;27:401–410. [Google Scholar]
- 29.Springer MS, Stanhope MJ, Madsen O, de Jong WW. Trends Ecol Evol. 2004;19:430–438. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Inagaki Y, Susko E, Fast NM, Roger AJ. Mol Biol Evol. 2004;21:1340–1349. doi: 10.1093/molbev/msh130. [DOI] [PubMed] [Google Scholar]
- 31.Lockhart PJ, Larkum AW, Steel M, Waddell PJ, Penny D. Proc Natl Acad Sci USA. 1996;93:1930–1934. doi: 10.1073/pnas.93.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown JH, Lomolino MV. Biogeography. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 33.Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M., FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 37.The Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 38.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. Science. 2007;316:222–234. [Google Scholar]
- 39.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 40.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 41.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, III, Zody MC, et al. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 42.Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 43.International Chicken Genome Sequencing Consortium. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Dudley J, Nei M, Kumar S. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 45.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony* (and Other Methods) Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 46.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 47.Guindon S, Gascuel O. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 48.Hordijk W, Gascuel O. Bioinformatics. 2005;21:4338–4347. doi: 10.1093/bioinformatics/bti713. [DOI] [PubMed] [Google Scholar]
- 49.Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 50.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 51.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 52.Pagel M, Meade A. Syst Biol. 2004;53:571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- 53.Cox CB, Healey IN, Moore PD. Biogeography: An Ecological and Evolutionary Approach. New York: Wiley; 1976. [Google Scholar]
- 54.Liu G, Uddin M, Islam M, Goodman M, Grossman LI, Romero R, Wildman DE. Source Code Med Biol. 2007 doi: 10.1186/1751-0473-2-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.