Abstract
Caspases are proteolytic enzymes that are essential for apoptosis. Understanding the many discrete and interacting signaling pathways mediated by caspases requires the identification of the natural substrate repertoire for each caspase of interest. Using an amplification-based protein selection technique called mRNA display, we developed a high-throughput screen platform for caspase family member specific substrates on a proteome-wide scale. A large number of both known and previously uncharacterized caspase-3 substrates were identified from the human proteome. The proteolytic features of these selected substrates, including their cleavage sites and specificities, were characterized. Substrates that were cleaved only by caspase-8 or granzyme B but not by caspase-3, were readily selected. The method can be widely applied for efficient and systematic identification of the family member specific natural substrate repertoire of any caspase in an organism of interest, in addition to that of numerous other proteases with high specificity.
Keywords: in vitro protein selection, mRNA display, natural caspase substrate repertoire, natural granzyme B substrates
Highly specific proteolysis of bioactive molecules is one of the most important mechanisms to achieve precise cellular control of many essential biological processes. Thus far, >500 genes that encode proteases or protease-like molecules have been annotated in the human genome (1). One of the most vital biochemical pathways that are mediated by proteases is apoptosis. Because caspase activation is a crucial event in apoptosis, identification of their downstream substrates on a proteome-wide scale is essential to our understanding of the molecular mechanisms of programmed cell death. Although a number of caspase substrates have been identified, it appears that many more apoptotic caspase targets, especially those caspase family member specific substrates, have not yet been revealed (2–4). The identification of family member specific substrates of each caspase on a proteome-wide scale, however, is a major undertaking. This challenge is further complicated by the overlapping substrate specificities of multiple caspase family members.
Four global methods have been used to identify the downstream substrates of caspases. The first method is a mass-spectrometric approach that permits identification of the cleavage sites of relatively abundant proteins under physiological conditions. However, it appears that this method predominantly identified some classes of substrates such as RNA-binding proteins (5). It is also difficult to unambiguously attribute a particular cleavage event to a specific caspase, because of the presence of multiple caspases and numerous other proteases in cell lysate. The second method is the small pool expression cloning strategy that has been successfully used to identify several previously uncharacterized caspase and recently granzyme B substrates (3, 6). It is time consuming to screen the whole cDNA library by progressive subdivision and reexamination of a large number of subpools each with hundreds of individual clones. The third method is yeast two-hybrid approach that uses inactive caspase mutants (7). This indirect binding rather than cleavage method would result in preferential enrichment of substrates that tightly bind to the caspase used in the screen. The fourth approach is to use a synthetic peptide library that is appropriately displayed, e.g., on phage, the bacterial cell membrane, or fluorogenic peptide microarrays. This approach has been successfully used to determine the specificity of various proteases including caspases (8, 9). However, the optimal consensus sequences identified from the short peptide libraries were poorly correlated with naturally occurring protease substrates and are therefore less biologically relevant. Identification of natural protease substrates by using these display methods has not yet been demonstrated. In general, these methods only permit the identification of a limited number of caspase substrates.
To identify natural family member specific caspase substrates on a proteome-wide scale, a systematic and broadly applicable strategy is greatly desired. mRNA display is a technology that allows covalent linkage between an RNA and its encoded protein (10–12). Using an mRNA-displayed protein library, multiple rounds of selection and amplification can be performed, enabling enrichment of rare sequences with desired properties. Compared with other peptide or protein selection methods, mRNA display has several advantages. First, the stable linkage between mRNA and protein makes it possible to use arbitrary selection conditions such as sequential cleavage with a series of different caspases. Therefore, it allows us to identify protein sequences that are cleaved only by the caspase member of interest, but not by other members. Second, protein libraries containing as many as 1012∼1013 unique sequences can be generated and selected, orders of magnitude higher than can be achieved by using other methods. Therefore, both the likelihood of isolating rare sequences and the diversity of sequences isolated in a given selection are significantly increased. Using mRNA-display-based protein selection, we identified and characterized a large number of caspase-3 substrates, including 26 known and 89 previously uncharacterized downstream caspase-3 targets [supporting information (SI) Tables 2–4]. The cleavage sites on the selected fragments from known caspase-3 substrates correlate very well with those previously reported and the cleavage sites on the previously uncharacterized substrates could be readily mapped. We also demonstrated that downstream substrates that were cleaved only by caspase-8 or granzyme B, but not by caspase-3, could be readily selected. These results will contribute to deciphering the apoptotic signaling pathways mediated by individual caspases or caspase-like proteases.
Results and Discussion
Natural Proteome Domain Library Used in the Proteolytic Selection.
The natural proteome domain library used in the selection was constructed with a mixture of Poly(A)+ mRNAs from human brain, heart, spleen, thymus, and muscle, as reported (12). Because the initial library was preselected to remove out-of-frame sequences, the percentage of the sequences in alternate frames was ≈3% and <1% before and after the proteolytic selection, respectively. For most sequences, the length of the protein portion from the ORFs of natural proteins was in the range of 80–200 aa. Protein domains with this length are more likely than short synthetic peptides to adopt native conformations or tertiary structures. Therefore, this method makes it possible to identify other unknown residues or structural motifs, in addition to the cleavage sites, which might play an important role in protease substrate recognition. Typically, 1.5 pmol (≈1012 molecules) of mRNA-displayed protein sequences was generated and used in the proteolytic selection. Therefore, each protein and its fragments should be represented by numerous copies in the library.
Selection Scheme for Efficient Enrichment of Substrates Cleaved by the Caspase of Interest.
To select caspase substrates from an mRNA-displayed human proteome domain library, we immobilized the proteome library on streptavidin-agarose beads through a biotin residue that was efficiently and site-specifically introduced at the N-terminal AviTag by using the BirA enzyme (13) (SI Fig. 6). Consequently, only sequences specifically cleaved by the caspase of interest were released and enriched. Several procedures were used to minimize the enrichment of nonspecific sequences, as detailed in Materials and Methods. Upon incubation with a purified caspase of interest, cleaved protein sequences were released and enriched, with the intact mRNAs still covalently attached to their C termini (Fig. 1). To avoid overdigestion, we first tested the in vitro digestion conditions for several well known caspase substrates, including PARP-1, calpastatin, RAD21, hnRNP A2/B1, and Bid. The incubation time and the amount of caspase used in the selection were chosen to result in ≈50% cleavage of these known substrates. The selected sequences were then amplified for sequencing or used as templates for iterative rounds of selection.
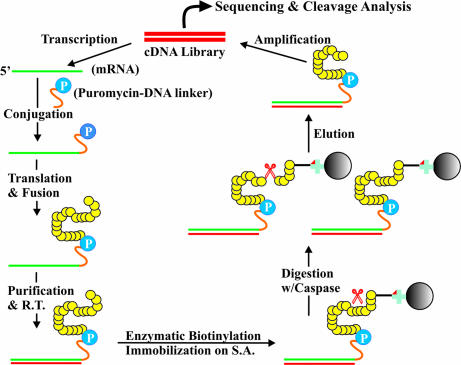
Fig. 1.
Selection scheme of caspase substrates using an mRNA-displayed human proteome domain library. mRNA, green; DNA, red; protein, yellow; puromycin, blue circle labeled as P; biotin, red triangle; streptavidin, blue cross; agarose beads, solid black circle; caspase, red scissors.
SI Fig. 7 shows the selection profiles using caspase-3. In the first two rounds, the amount of immobilized fusion molecules released from the solid surface by caspase-3 was undetectable (<0.1%). In round 3, however, ≈1.5% of total immobilized radioactive counts were recovered. By cloning and sequencing, it was found that 99% of the selected sequences were in the correct reading frame as in the natural proteins. The selected sequences were diverse, although fragments from some genes had more copies than others. To facilitate the isolation of additional substrate sequences, 12 most-abundant sequences were removed by hybridizing the selected pool with 18 biotinylated oligonucleotides complementary to the shortest overlapping regions of each abundant sequence (SI Tables 5 and 6), followed by passage through a streptavidin-agarose column. It was found that such subtraction was very efficient, and the abundance of these sequences was significantly reduced by only one such subtraction (SI Table 5). After this treatment, a large number of previously uncharacterized caspase-3 substrates, which otherwise might be difficult to detect, were successfully identified.
Efficient Proteolytic Analysis of Selected Sequences.
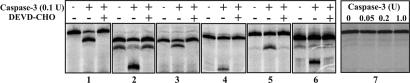
The cleavage of each selected protein fragment by caspase-3 was tested by an in vitro proteolytic assay, using radiolabeled protein generated by coupled in vitro transcription and translation (TNT), in the presence or absence of a caspase-3 inhibitor (Ac-DEVD-CHO). Each reaction mixture was loaded to a high resolution Tricine-SDS/PAGE for analysis. Numerous positive and negative controls were tested to confirm that this in vitro proteolytic assay is robust, provided that the protein of interest is translated well in rabbit reticulocyte lysate (data not shown). As illustrated in Fig. 2 using six selected proteins as examples, the cleavage of positive sequences was observed when purified caspase-3 was present, but totally abolished when caspase-3 was preinhibited by a specific inhibitor. Among 207 unique proteins from a total of 580 clones we picked up for analysis, 173 were translated well by TNT, among which 115 proteins (66%) were found to be specifically cleaved by caspase-3. These 115 caspase substrates are listed in Table 1 and SI Table 2 for new and previously characterized caspase-3 substrates, respectively. Most of the selected sequences contain 80 to 200 aa residues from the corresponding ORFs. A typical selected sequence is shown in Supplemental Fig. 3. Blast analysis indicates that this is a fragment (Gln32-Gly154) from the N terminus of protein kinase C-like 2 (PKN2, full-length 984-residue), a caspase-3 substrate previously identified using the small pool expression cloning strategy (3).
Fig. 2.
In vitro proteolytic cleavage of several protein fragments isolated from round 3 selection using caspase-3. Radiolabeled protein fragments were synthesized by TNT. The cleavage was carried out using 0.1 unit/μl purified caspase-3 at 37°C for 2 h. In the inhibition assay, caspase-3 was preinhibited by 0.5 μM Ac-DEVD-CHO for 15 min before being used to digest TNT products. Different proteins were not aligned according to their molecular weights. 1 is a positive control fragment originated from a cleavage site containing sequence of PARP-1. 2–6 are fragments from nos. 4, 5, 27, 35, and 47, as listed in SI Table 4. 7 is a negative control sequence randomly picked up from round 0. This sequence was digested with increased amount of caspase-3 to demonstrate that its noncleavage nature was not changed by using 10-fold higher caspase.
Table 1.
Previously uncharacterized caspase-3 substrates isolated from round 3 of selection
| Number | Protein name | Selected SOR/full length | Tested FL or LF* | Cut by caspase-3? |
|---|---|---|---|---|
| 1 | ADNP | 824–929/1102 | ||
| 2 | AFTIPHILIN | 108–208/908 | ||
| 3 | AGGF1 | 504–592/714 | 442–714 | Y |
| 4 | ALS2C13 | 120–223/345 | ||
| 5 | ANKRD1 | 26–97/319 | 1–319 | Y |
| 6 | ARGAP21 | 1400–1498/1957 | ||
| 7 | ARGEF12 | 683–776/1544 | ||
| 8 | ARL13B | 304–368/428 | ||
| 9 | BCAS1 | 244–301/584 | 1–584 | Y |
| 10 | BNIP2 | 4–79/314 | 1–314 | Y |
| 11 | BTAF1 | 124–241/1849 | 1–472 | Y |
| 12 | BRWD3 | 1588–1641/1802 | 688–1384 | Y |
| 13 | C10ORF6 | 336–446/1173 | ||
| 14 | C12ORF11 | 585–696/706 | ||
| 15 | C219 | 599–711/1907 | 493–974 | Y |
| 16 | CCDC47 | 79–192/483 | 1–483 | Y |
| 17 | CDC10 | 92–240/417 | 1–417 | N |
| 18 | CEP1 | 1333–1438/2325 | ||
| 19 | CGNL1 | 630–725/1302 | 446–912 | Y |
| 20 | CHD3 | 602–696/1966 | ||
| 21 | CLU | 116–213/501 | 53–501 | Y |
| 22 | COG5 | 450–575/839 | ||
| 23 | DGKB | 354–453/773 | ||
| 24 | DHX38 | 7–99/1227 | 1–674 | Y |
| 25 | DNM1 | 547–647/851 | ||
| 26 | DYN | 2738–2830/5171 3384–3480/5171 | ||
| 27 | EEF1B2 | 59–168/225 | 1–225 | N |
| 28 | EPRS | 921–1022/1512 | 693–1315 | N |
| 29 | ESRRBL1/HIPPI | 275–389/429 | 1–429 | Y |
| 30 | FILIP1 | 709–848/1213 | ||
| 31 | FLJ21908 | 351–457/665 | ||
| 32 | FUS | 273–357/526 | ||
| 33 | GAPVD1 | 1051–1163/1487 | 940–1487 | Y |
| 34 | GOLGA4 | 681–819/2230 | ||
| 35 | GOLGB1 | 1833–1978/3259 | 1095–1701 | Y |
| 36 | HSPA4 | 640–784/840 | 1–840 | N |
| 37 | HYPB | 781–900/2061 | 688–1263 | Y |
| 38 | INO80 | 1322–1391/1556 | 1207–1556 | Y |
| 39 | KIF21A | 558–655/1661 | ||
| 40 | JARID1A | 1589–1685/1690 | 1131–1690 | Y |
| 41 | KIAA0423 | 871–974/1720 | 593–1180 | Y |
| 42 | KIAA0864 | 159–263/1402 | ||
| 43 | KIDIN220 | 221–301/1771 | 1–627 | Y |
| 44 | LMO7 | 1085–1184/1349 | 553–1271 | Y |
| 45 | LOC22998 | 684–803/1083 | ||
| 46 | M11S1 | 80–165/709 | 54–709 | Y |
| 47 | MACF1 | 1661–1757/5430 4126–4240/5430 | 1427–2062 | Y |
| 48 | MATRIN3 | 636–739/847 | ||
| 49 | MLL3 | 810–910/4025 | 560–1126 | Y |
| 50 | MNAB | 1033-1113/1191 | 652-1191 | Y |
| 51 | MORF15 | 145–292/362 | 1–362 | N |
| 52 | MPP8 | 432–513/860 | ||
| 53 | MYO18A | 1943–2047/2054 | ||
| 54 | NAPLl3 | 97–181/506 | 1–506 | Y |
| 55 | NDUFS1 | 240–328/727 | 1–727 | Y |
| 56 | NEB | 832–928/6669 | 550–1058 | Y |
| 57 | NEXN | 309–355/505 | 1–505 | Y |
| 58 | NUCB2 | 175–269/420 | 1–420 | Y |
| 59 | PCF11 | 319–442/1555 | 1–713 | Y |
| 60 | PDZGEF1 | 329–439/1499 | 130–950 | Y |
| 61 | PHACTR2 | 511–592/634 | 1–634 | Y |
| 62 | PHTF2 | 267–365/785 | ||
| 63 | PIK3R1 | 501–631/724 | ||
| 64 | PIK3R3 | 64–215/461 | 1–461 | Y |
| 65 | PJA2 | 131–237/708 | 1–708 | Y |
| 66 | PPP1R12A | 833–916/1030 | ||
| 67 | PSIP2 | 69–148/530 | 1–530 | Y |
| 68 | PSMC1 | 251–323/440 | 1–440 | |
| 69 | PSMC3 | 269–348/439 | 1–439 | |
| 70 | PSRC2 | 317–395/1989 | ||
| 71 | RNF6 | 128–227/685 | 1–685 | Y |
| 72 | S100PBP | 85–188/408 | ||
| 73 | SALL1 | 97–134/1324 | ||
| 74 | SCG3 | 78–138/468 | 1–468 | Y |
| 75 | SERBP1 | 180–256/402 | ||
| 76 | SHOC2 | 191–308/582 | 1–582 | Y |
| 77 | SLC4A1AP | 711–782/796 | 1–796 | Y |
| 78 | SMARCA1 | 856–923/1054 | ||
| 79 | SMC5 | 807–910/1101 | ||
| 80 | SORBS1 | 389–525/816 | 1–816 | Y |
| 81 | SPARCL1 | 257–335/664 | 1–664 | Y |
| 82 | SSFA2 | 439–534/1256 | ||
| 83 | SRP54 | 358–436/504 | 1–504 | Y |
| 84 | STAC3 | 259–363/364 | 1–364 | N |
| 85 | SYTL2 | 686–797/1256 | ||
| 86 | TAF15 | 99–171/589 | ||
| 87 | TFG | 23–145/400 | 1–400 | Y |
| 88 | UTP11L | 109–196/253 | 1–253 | N |
| 89 | UTP14C | 305–395/766 | 1–766 | Y |
Only selected fragments whose specific cleavage by caspase-3 has been confirmed by SDS/PAGE are listed. Column 3 shows the range of the shortest overlap region (SOR) and the size of the full-length protein. Column 4 shows the range of the TNT-generated full length (FL) or of large fragment (LF) that had been tested in caspase-3 cleavage reaction (the results are listed in column 5). The GenBank accession numbers of these proteins are listed in SI Table 4. Y, yes; N, no. Bold text represents full-length proteins that were specifically cleaved by caspase-3, whereas italicized text represents large fragments from very large proteins that were specifically cleaved by caspase-3.
*Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan) and Protein Blast were used for the prediction of domains, motifs, and patterns. The Hierarchical Neural Network method (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_nn.html) was used for the prediction of secondary structures.
Among these 115 positive sequences, 26 were fragments from previously reported caspase-3 substrates (SI Tables 2 and 3). Unlike other methods that result in the enrichment of certain classes of proteins (14), the caspase-3 substrates selected here include regulatory proteins involved in various signaling pathways and are not limited to any particular class.
We were not able to identify all of the caspase-3 substrates reported in the literature. However, the absence of those known caspase-3 substrates from our list does not necessarily mean they were not selected, because rare sequences are less likely to be picked up for follow-up studies. We hypothesized that short fragments surrounding the cleavage sites should be present, if these proteins were enriched in the selection. We examined 18 known caspase-3 substrates that were not found from the clones we randomly picked up for analysis. The presence of these genes in the selected pool was detected by PCR amplification using gene-specific primers that target short fragments surrounding the reported caspase-3 cleavage sites. 12 of 18 such substrates were greatly enriched in the selected pool, as they could be amplified to the same levels as the controls by at least 12 fewer PCR cycles. Because such PCR detection approach is only limited to known caspase substrates whose cleavage sites had been characterized, we focused our efforts on studying previously uncharacterized caspase-3 substrates.
Mapping of the Potential Cleavage Sites on the Selected Substrates.
The ease of mapping potential cleavage sites is one of the biggest advantages of this method. As shown in SI Tables 2 and 3, almost all of the selected fragments from the known caspase-3 substrates contain the previously reported cleavage site(s) (4), clearly indicating that our method allows efficient identification of caspase substrates on a proteome-wide scale. Our results also allow us to assign putative cleavage sites to these known caspase-3 substrates whose cleavage sites have not yet been characterized. One such example is RANBP2, a nucleoporin that was recently found to act as an E3 ligase by binding both SUMO and Ubc9 to position the SUMO-E2-thioester in an optimal orientation to enhance conjugation (15). By analyzing the fragments we isolated from RANBP2, it was found that potential cleavage sites could be located at DVTD111/G, DFAD2411/G, or DVAD2430/S. These cleavages will remove the tetratricopeptide repeat (TPR) domain (one TPR, residues 60–93) from the N terminus if the cleavage occurs at D111 or truncate the C-terminal cyclophilin ABH-like domain if the cleavage is at D2411 or D2430. We were not able to overexpress RANBP2 in 293 cells because of its very large size. Nevertheless, our result suggests that the functions of RANBP2 could be regulated by caspase-3 during apoptosis through cleavage of these domains. Of the fragments selected from 89 previously uncharacterized caspase-3 substrates whose cleavage was individually confirmed by in vitro proteolytic assay, most contain only one potential DXXD cleavage site (SI Table 4). The potential caspase-3 cleavage on such fragments most likely occurs on these residues. For the few selected fragments that contain more than one potential caspase-3 cleavage site, we narrowed down the cleavage site(s) by mapping the shortest overlapping region. This is possible, because a number of sequences were isolated as multiple fragments of different lengths derived from the same parental ORF. For example, by aligning three PKN2 fragments isolated from the selection, the shortest overlapping region was found to be between residues 65–140, which covers only DEPDITD117/C but not the other two potential cleavage sites (SI Fig. 8). This DEPDITD117/C cleavage site was further confirmed by analyzing the sizes of the proteolytic products using high resolution Tricine-SDS/PAGE (data not shown). Indeed, the in vitro cleavage site thus identified is identical to that mapped under physiological conditions (3). However, it is still possible that in some cases more than one potential cleavage site is present in the shortest overlapping region, as in secretogranin III (SCG3), an acidic secretory granin protein that is specifically expressed in neuronal and endocrine cells. All of the isolated protein fragments originating from SCG3 contain a common region from residues Leu78 to Leu138, which includes the two potential cleavage sites DDYD121/S and DDPD136/G. In vitro proteolytic analysis using in vitro-translated fragment and full-length SCG3 showed that the cleavage sites were indeed within this common region. Because these two potential cleavage sites are very close to each other, it is difficult to unambiguously determine which one is cleaved using Tricine-SDS/PAGE. Nevertheless, this approach allows us to effectively narrow down the potential cleavage site(s) to a region of <20 residues.
Proteolytic Analysis of Full-Length Proteins.
We further investigated the caspase-3-mediated cleavage of those proteins whose full-length form could be expressed well using the TNT system. Of 33 such full-length proteins from the 115 that were shown to be cleavable as fragments, 26 (≈79%) were specifically cleaved by caspase-3 (Table 1, column 4, bolded). Interestingly, a number of the selected fragments were from very large proteins with >1,000 aa that we could not express as full-length proteins. For these proteins, we generated large fragments of 500–800 aa that cover the selected sequences and used these large fragments to test cleavage by caspase-3. The boundaries of these fragments were chosen on the basis of online domain prediction algorithms Motif Scan and Hierarchical Neural Network (Table 1 and SI Table 4). Interestingly, 19 of 20 such selected sequence-containing large fragments were cleaved by caspase-3 (Table 1, column 4, italicized). The sizes of the observed cleavage fragments on SDS/PAGE were consistent with those predicted based on the mapped cleavage sites. These results demonstrate that although the caspase-3 cleavable sequences isolated from the selection were relatively short (80–200 residues), most of the corresponding full-length or large fragments were indeed cleaved by caspase-3.
Caspase Specificity of Selected Substrates.
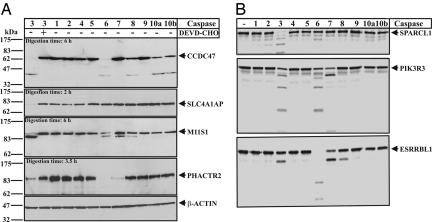
Because of the presence of multiple caspases in eukaryotic cells, it is of great importance to determine which member of the caspase family is responsible for the observed cleavage. To address this issue, we investigated the cleavage specificity of several potential previously uncharacterized caspase-3 substrates with 10 human caspase members (Fig. 3). These full-length proteins were either overexpressed in HEK 293T (Fig. 3A) or generated by TNT in rabbit reticulocyte lysate (Fig. 3B). We found that affinity-tagged overexpressed proteins listed in Fig. 3A were cleaved during apoptosis, but the cleavage was not blocked by caspase inhibitors, possibly because they were also cleaved by other enzymes that were present in cells. Therefore, we added purified caspase family members to uninduced cell lysates to investigate whether the overexpressed proteins in mammalian cells were indeed cleaved by caspases (Fig. 3A). Consistent with the conditions applied in the selection, all of these proteins could be efficiently cleaved by caspase-3 but not by caspase-3 preincubated with a specific tetrapeptide inhibitor. The cleavages were all time-dependent, as shown in SI Fig. 9 or PIK3R3 as an example. Interestingly, most of these proteins were also digested by caspase-6 and/or -7. The cleavage patterns are very similar to that by caspase-3 but could be different as shown in ESRRBL1 (Fig. 3B). This is presumably because caspases-3, -6, and -7 belong to the same family of executioner caspases and therefore have similar specificities. However, the extent and/or pattern of cleavage by other executioner caspases (-6 and -7) could be different, suggesting they have different cleavage efficiencies or recognition sites. Interestingly, several substrates were also cleaved by caspase-8 with similar cleavage patterns. This result indicates that caspase-3 and -8 share some common substrates, although they have different cleavage specificities.
Fig. 3.
Caspase cleavage specificity of full-length proteins. (A) Ex vivo proteolytic cleavage of several full-length proteins by different human caspase members. The V5/His-tagged full-length proteins were overexpressed in HEK 293T cells. The cleavage was carried out using 0.1 unit/μl various purified caspases at 37°C for 6 h with an appropriate amount of cell lysate. The proteins were detected by Western blotting using anti-V5 antibody. In the inhibition assay of caspase-3, caspase-3 inhibitor Ac-DEVD-CHO was used at 10 μM. (B) Specificity of in vitro proteolytic cleavage of several radiolabeled full-length proteins generated by TNT. The cleavage was carried out by using 0.1 unit/μl of various purified caspases at 37°C for 3 h. The full names and the clone numbers of the proteins are listed in SI Table 4. The clone numbers of the proteins shown are: CCDC47 (no. 16), SLC4A1AP (no. 77), M11S1 (no. 46), PHACTR2 (no. 61), SPARCL1 (no. 81), PIK3R3 (no. 64), and ESRRBL1 (no. 29).
Substrates Cleaved Only by Caspase-8 Could Be Readily Selected.
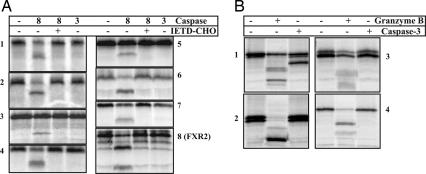
To demonstrate that our method is not limited to caspase-3 but can be generally used to identify downstream substrates of other members of caspase, we performed another proteome-wide selection using caspase-8. We then studied the cleavage of the proteins isolated from the caspase-8 selection by either caspase-3 or -8. More than 30 proteins were found to be cleaved by caspase-8 (unpublished data). Many of the selected caspase-8 substrates could also be cleaved by caspase-3, presumably because some caspase-8 cleavage sites are recognized by caspase-3. Significantly, a number of these selected caspase-8 substrates were specifically cleaved by caspase-8 but not by caspase-3 (Fig. 4A). These results clearly demonstrate that the mRNA display-based proteolytic selection can be used to decipher family member-specific substrates for each caspase in the caspase family.
Fig. 4.
Proteolytic cleavage of fragments from caspase-8 and granzyme B selections. (A) In vitro proteolytic cleavage of several protein fragments isolated from round 5 selection using caspase-8. Radiolabeled protein fragments were synthesized by TNT. The cleavage was carried out by using 0.1 unit/μl of purified caspase-3 or -8 at 37°C for 6 h. In the inhibition assay of caspase-8, caspase-8 inhibitor Ac-IETD-CHO was used at 5 μM. The proteins are: 1, PJA2; 2, ZCWPW2; 3, POLR3GL; 4, NRG1; 5, FLJ21352; 6, KIAA0701; 7, EEA1; and 8, FXR2. (B) In vitro proteolytic cleavage of several protein fragments isolated from round 2 selection using granzyme B. Radiolabeled protein fragments were synthesized by TNT. The cleavage was carried out by using 0.1 unit/μl of purified caspase-3 or 0.2 unit/μl granzyme B at 37°C for 6 h. The proteins are: 1, SERBP1; 2, KBTBD10; 3, C11ORF58; and 4, FILIP1L. The full names of these proteins are given in SI Table 6.
Caspase-3 and Granzyme-B Substrates Could Be Readily Distinguished.
We also used this method to study the substrates of granzymes, a class of specific proteases that play central roles in cytotoxic lymphocyte-mediated apoptosis. Like caspases, granzyme B is a specific protease with a stringent preference of an aspartic residue at the P1 position and therefore shares many substrates with caspases (8, 16, 17). To understand the apoptotic signaling pathways specifically mediated by granzyme B, it is of great importance to identify the substrates that are only cleaved by granzyme-B but not by caspases. It was reported that residues outside the P4-P2′ motif also contribute to the substrate recognition and binding by granzyme B, making the identification of granzyme B-specific substrates more challenging than that of caspases. We attempted to enrich such specific substrates of granzyme B by preremoval of sequences cleavable by caspases through incubating an immobilized, mRNA-displayed human proteome domain library with a mixture of caspases (caspases-2, -3, -6, -7, -8, -9, and -10) before selecting sequences that can be released by purified granzyme B. More than 60 granzyme B-substrates were successfully identified (unpublished data). Fig. 4B shows a few such examples. Only a few of the selected sequences, such as SERBP1, were still cleaved by caspases, but usually with different cleavage patterns. This preremoval of sequences that are recognized by other proteases can be widely used in such mRNA display-based method to identify substrates that are cleaved only by a protease of interest.
Cleavage of the Selected Substrates Under in Vivo Conditions.
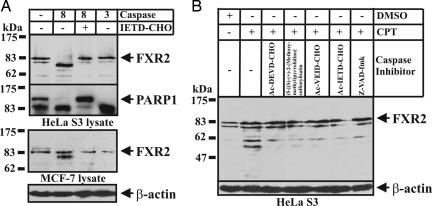
We also investigated whether the cleavage of several selected caspase substrates indeed occurs during apoptosis when the proteins are present at their endogenous concentrations. The in vivo cleavage of the known caspase-3 substrates listed in SI Table 2 has been reported by numerous studies (4). Because most previously uncharacterized substrates of interest do not have commercially available antibodies, it is difficult to investigate them in vivo when the proteins of interest are present at their endogenous concentrations. We first demonstrated that the cleavage of endogenous SRP54 and EIF4B were observed in vivo under apoptotic conditions (SI Fig. 10). We then examined the cleavage of fragile X mental retardation syndrome-related protein 2 (FXR2), a caspase substrate identified from the selection using caspase-8 rather than caspase-3. FXR2 is similar to FMRP and FXR1, both of which are involved in RNA binding, polyribosomal association, and nucleocytoplasmic shuttling (18). These genes have been implicated in the development of fragile X mental retardation syndrome. The shortest overlapping region on FXR2 recognized by caspase-8 was found to be between Leu564-Ser634, and the potential cleavage site was mapped at LESD567/G (SI Fig. 11). Interestingly, this cleavage site is just 13 residues upstream of the functional nucleolar-targeting signal (Arg581-Arg594), which is involved in mediating the intranuclear distribution of FXR2 and some FXR1 isoforms (18). We found that full-length FRX2 was specifically cleaved by caspase-8, but not by caspase-3, when digested using purified caspase-3 or -8, respectively (Fig. 5A). The sizes of the proteolytic products were consistent with a caspase-8 cleavage site at D567/G. Its in vivo cleavage under apoptotic conditions was also observed, although it is less clear whether the cleavage is mediated by only one caspase (Fig. 5B). It appears that the large fragment was further digested by other caspases or proteases in vivo. Our results suggest that the C-terminal nucleolar-targeting signal of FXR2 is removed by caspase during apoptosis, thus effectively preventing its localization in nucleolus and disrupting most of its functions.
Fig. 5.
Ex vivo and in vivo cleavage of FXR2 during CPT-mediated apoptosis. (A) Western blotting analysis of ex vivo cleavage of FXR2 by caspase-3 or -8. Caspase-8 or -3 (0.1 unit/μl) was added to HeLa S3 or MCF-7 whole-cell lysates for 6 h at 37°C, in the presence or absence of Ac-IETD-CHO at 5 μM. The treated lysate was separated on SDS/PAGE gels and probed with anti-FXR2. The ex vivo cleavage of PARP1 was monitored as a positive control. (B) Western blotting analysis of the in vivo cleavage of FXR2 during apoptosis in HeLa S3 cells induced by CPT. Cells were treated with 2 μM CPT for 12 h. For the FRX2 cleavage inhibition assay, cells were pretreated with a caspase inhibitor at 50 μM for 3 h before the addition of CPT. The blot membranes were probed with anti-FXR2.
Limitations and Advantages of the Method.
Although mRNA display is a powerful method that allows for efficient identification of caspase family member-specific substrates, this method does have its limitations. The efficiency with which a particular sequence is selected depends on several factors, including its mRNA abundance and the efficiencies of protein expression and caspase-catalyzed cleavage. This might explain why only a fraction of known caspase-3 substrates were identified. However, because we were not able to exhaustively analyze the selected pool, more known substrates could be identified by analyzing more colonies. In addition, the substrates reported here were identified and characterized by in vitro approaches using purified caspases. Whether a specific protein is cleaved during apoptosis under physiological conditions requires individual examination. Therefore, this method is complementary to previously used methodologies. Nevertheless, this method is thus far the most broadly applicable strategy for the identification of a large number of diverse caspase substrates. The in vitro nature of this method allows us not only to attribute the cleavages to a specific caspase member but also to efficiently remove the abundant or biased sequences. The method reported here is systematic and high throughput and can be broadly applied to all members of human caspases, caspases in other organisms, and numerous other proteases with high specificity. Therefore, the natural substrate repertoire of caspases in other organisms, including CED-3, DRONC, and DrICE from Caenorhabditis elegans and Drosophila, respectively, should be readily identifiable using the same approach. The availability of this information will greatly facilitate our understanding of apoptotic signaling pathways.
Materials and Methods
Generation of the mRNA-Displayed Proteome Domain Library and Site-Specific Biotinylation of the N Termini of the Protein Sequences.
The library was constructed as detailed in SI Text. A sequence coding an AviTag was introduced at the very N terminus of the ORF (13). mRNA-protein fusions were generated and purified from the lysate, as described (12), followed by incubation with an appropriate amount of BirA (Avidity, Denver, CO) in a biotinylation buffer at 30°C for 4 h for site-specific in vitro biotinylation. After reverse transcription to convert the fusion molecules to DNA/RNA hybrids, the mRNA-displayed proteome domain library was purified on the basis of the affinity tags.
Selection of Caspase Substrates from mRNA-Displayed Human Proteome Domain Library.
The purified proteome domain library (≈1.5 pmol each round) was first diluted in a caspase reaction buffer with 10 mM DTT and 1 mM EDTA and passed through a precolumn of 100 μl of streptavidin-agarose beads preblocked with excess amount of biotin. The flowthrough was then incubated with the same amount of streptavidin-agarose beads for 1 h at room temperature. Unbound and nonspecifically bound molecules were washed off the column using 250 column volumes of reaction buffer. To further remove sequences that might be nonproteolytically released, we carried out a preincubation under the otherwise same reaction conditions, except BSA was used to replace the caspase of interest. The immobilized proteome domain library was then incubated with appropriate amount of purified caspase (BioVision, Mountain View, CA) in 300 μl of reaction mixture at 37°C for 1.5 h with rotation. Proteins specifically cleaved by the caspase of interest were released from the solid surface. The selected molecules were PCR-amplified for the next round of selection or cloned into a TOPO vector for sequencing and analysis.
In Vitro Proteolytic Analysis of Selected Substrates.
Individually cloned sequences isolated from the selection were PCR-amplified using two consensus primers. Full-length genes of interest were RT-PCR-amplified using gene-specific primers from a human cDNA library. Purified PCR products were used as templates for TNT in the presence of 10 μCi (1 Ci = 37 GBq) [35S]methionine in a total volume of 12.5 μl for 90 min at 30°C. To perform in vitro proteolytic analysis, an aliquot of each translation reaction mixture was incubated in a reaction buffer at 37°C for 2–6 h with an appropriate amount of active caspase or caspase preinhibited by a tetrapeptide inhibitor. An aliquot of each reaction mixture was loaded onto an SDS/PAGE gel for separation and signal detection.
Ex Vivo Cleavage Assay.
Caspase-catalyzed proteolysis of overexpressed proteins was performed by the addition of an appropriate amount of exogenous caspase to HEK 293T whole-cell extracts. Each digestion reaction contains 0.2–2 units of caspase and 25–50 μg of total protein from cell lysate. The reaction mixture was incubated at 37°C for various times. Similarly, cleavage analysis of endogenous FXR2 was accomplished by adding caspase-8 or -3 to the cleared untreated HeLa S3 or MCF-7 lysates. Inhibition reactions were performed by preincubating 5 μM caspase-3 (Ac-DEVD-CHO) or caspase-8 (Ac-IETD-CHO) inhibitor with the corresponding caspase for 30 min at 37°C. Afterward, 25–50 μg of cell lysate was added and incubated for an additional 6 h at 37°C. Proteins in lysates were resolved by SDS/PAGE, and blots were probed with anti-V5 (Invitrogen), anti-FXR2 (Abcam, Cambridge, MA), or anti-PARP1 (Upstate Biotechnology, Billerica, MA) as a positive control.
In Vivo Cleavage Assay.
For caspase-catalyzed in vivo cleavage of BID, EIF4B, SRP54, and FXR2, 2 μM camptothecin (CPT) (Calbiochem, San Diego, CA) was directly added to growing HeLa S3 to induce apoptosis. For cleavage inhibition assay, cells were pretreated with 50 μM of a particular caspase inhibitor for 3 h at 37°C, before induction of apoptosis by CPT. Approximately 25–50 μg of cell lysate was loaded into each well, separated by SDS/PAGE. Blots were probed with various antibodies including anti-BID (BioVision), anti-EIF4B (Cell Signaling Technology, Danvers, MA), anti-SRP54 (BD Biosciences, San Jose, CA), anti-FXR2, and anti-β actin (Sigma, St. Louis, MO).
For further information, see SI Figs. 6–11 and Tables 2–6.
Supplementary Material
Acknowledgments
This work was supported by startup funds from the Carolina Center for Genome Sciences and School of Pharmacy at the University of North Carolina at Chapel Hill and National Institutes of Health Grant NS047650 (to R.L.).
Abbreviation
- CPT
camptothecin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702251104/DC1.
References
- 1.Lopez-Otin C, Overall CM. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 2.Stroh C, Schulze-Osthoff K. Cell Death Differ. 1998;5:997–1000. doi: 10.1038/sj.cdd.4400451. [DOI] [PubMed] [Google Scholar]
- 3.Cryns VL, Byun Y, Rana A, Mellor H, Lustig KD, Ghanem L, Parker PJ, Kirschner MW, Yuan J. J Biol Chem. 1997;272:29449–29453. doi: 10.1074/jbc.272.47.29449. [DOI] [PubMed] [Google Scholar]
- 4.Fischer U, Janicke RU, Schulze-Osthoff K. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockstedt E, Rickers A, Kostka S, Laubersheimer A, Dorken B, Wittmann-Liebold B, Bommert K, Otto A. J Biol Chem. 1998;273:28057–28064. doi: 10.1074/jbc.273.43.28057. [DOI] [PubMed] [Google Scholar]
- 6.Loeb CR, Harris JL, Craik CS. J Biol Chem. 2006;281:28326–28335. doi: 10.1074/jbc.M604544200. [DOI] [PubMed] [Google Scholar]
- 7.Kamada S, Kusano H, Fujita H, Ohtsu M, Koya RC, Kuzumaki N, Tsujimoto Y. Proc Natl Acad Sci USA. 1998;95:8532–8537. doi: 10.1073/pnas.95.15.8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Proc Natl Acad Sci USA. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulware KT, Daugherty PS. Proc Natl Acad Sci USA. 2006;103:7583–7588. doi: 10.1073/pnas.0511108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts RW, Szostak JW. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu R, Barrick JE, Szostak JW, Roberts RW. Methods Enzymol. 2000;318:268–293. doi: 10.1016/s0076-6879(00)18058-9. [DOI] [PubMed] [Google Scholar]
- 12.Shen X, Valencia CA, Szostak J, Dong B, Liu R. Proc Natl Acad Sci USA. 2005;102:5969–5974. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckett D, Kovaleva E, Schatz PJ. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiede B, Dimmler C, Siejak F, Rudel T. J Biol Chem. 2001;276:26044–26050. doi: 10.1074/jbc.M101062200. [DOI] [PubMed] [Google Scholar]
- 15.Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT. Nat Struct Mol Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 16.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 17.Kam CM, Hudig D, Powers JC. Biochim Biophys Acta. 2000;1477:307–323. doi: 10.1016/s0167-4838(99)00282-4. [DOI] [PubMed] [Google Scholar]
- 18.Tamanini F, Kirkpatrick LL, Schonkeren J, van Unen L, Bontekoe C, Bakker C, Nelson DL, Galjaard H, Oostra BA, Hoogeveen AT. Hum Mol Genet. 2000;9:1487–1493. doi: 10.1093/hmg/9.10.1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.