Abstract
The yeast repressor Rme1p acts from distant binding sites to block transcription of the chromosomal IME1 gene. Rme1p can also repress the heterologous CYC1 promoter when Rme1p binding sites are placed 250–300 bp upstream of CYC1 transcriptional activator binding sites (UAS1 and UAS2). Here, in vivo footprinting studies indicate that Rme1p acts over this distance by preventing the binding of the CYC1 transcriptional activators to UAS1 and UAS2. Inhibition of activator binding by Rme1p has the same genetic requirements as repression: both depend upon sequences flanking the Rme1p binding sites and upon Rgr1p and Sin4p, two subunits of the RNA polymerase II-associated Mediator complex that are required for normal nucleosome density. Thus Rme1p may alter chromatin to prevent binding of transcriptional activators to distant DNA sequences.
Eukaryotic transcriptional repressors act through a variety of mechanisms to inhibit gene expression. Some repressors compete for DNA binding sites with specific activators; others block activity of particular transcriptional activators or interfere more directly with global factors required for transcriptional initiation (reviewed in refs. 1–3). A unique repression mechanism, transcriptional silencing, is used to repress specific chromosomal regions in yeast (reviewed in refs. 4 and 5). Silencing results from a change in chromatin structure that prevents interaction between DNA and DNA binding proteins, such as endonucleases, Dam methylase, and, by inference, transcription factors. Silencing in yeast results in permanent repression, whereas other repression mechanisms are generally used transiently, to permit gene expression levels to respond to genetic or environmental signals. Our studies reported here indicate that a yeast repressor, Rme1p, uses a mechanism akin to silencing to achieve regulated repression.
Rme1p (Regulator of Meiosis) is an inhibitor of meiosis and subsequent spore formation (refs. 6 and 7; reviewed in refs. 8 and 9). RME1 is expressed in a and α cells, which are unable to enter meiosis, and is repressed in a/α cells, which can enter meiosis (10). Rme1p blocks meiosis by preventing expression of IME1 (11), a positive regulator of many meiosis-specific genes (reviewed in ref. 9). Repression occurs only in starved a and α cells (11, 12), when Rme1p expression increases 10-fold over the level in growing cells (13, 14). Rme1p expression may increase after starvation as a consequence of G1 arrest (15). Rme1p represses IME1 directly, because repression depends upon two Rme1p binding sites in the IME1 5′ regulatory region (ref. 13; M.S., W.L., P. A. Covitz, H.S., and A.P.M., unpublished work).
Repression by Rme1p is unusual in that it is exerted over a considerable distance. The Rme1p binding sites lie over 1600 bp upstream of IME1 RNA start sites and over 600 bp upstream of the IME1 upstream activation sequence (UAS) region (refs. 16 and 17; see Fig. 1A). Other yeast repressors generally act over smaller distances in chromosomal promoter regions (18–20). Repression depends upon both Rme1p binding sites and upon flanking chromosomal sequences. A minimal region for repression has been defined through insertions of IME1 sequences upstream of a CYC1-lacZ reporter gene (13). This region, referred to as the Repression Cassette (RC), includes Rme1p binding sites and sequences between −1873 and −1743 from the IME1 locus (ref. 13; see Fig. 1B). Indeed, Rme1p is a weak transcriptional activator in the absence of the −1873/−1743 region (13, 15). The ability of Rme1p to activate transcription in isolation and the size of the region required for repression distinguish this repression system from many others.
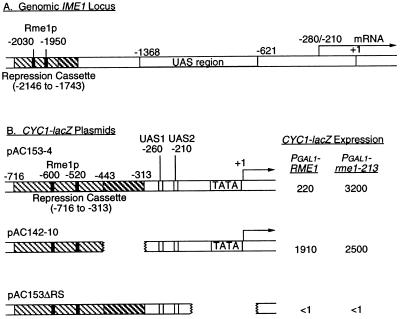
Figure 1.
Structure of the IME1 locus and CYC1-lacZ derivatives. (A) The genomic IME1 locus, indicating Rme1p binding sites (at positions −1950 and −2030; refs. 4 and 5), the RC (positions −1743 to −2146; hatched boxes), the UAS region (positions −1368 to −621; ref. 6), and RNA start sites (within the −210 to −280 interval; refs. 27 and 28). (B) CYC1-lacZ reporter genes, present on high-copy plasmids. Locations of the RC, UAS1, UAS2, TATA region, and RNA start sites are diagrammed. CYC1-lacZ expression is indicated (in Miller units of β-galactosidase) for strains expressing functional Rme1p (PGAL1-RME1) and nonfunctional Rme1–213p (PGAL1-rme1–213).
Genetic studies (21) have established a connection between Rme1p and the Rgr1p–Sin4p complex (22), which seems to have two distinct roles in transcriptional control. One set of observations suggests that Rgr1p and Sin4p have a role in maintenance of chromatin structure. In rgr1 and sin4 mutants, negative superhelicity of plasmids is decreased, as might arise from a decrease in nucleosome density (22). In addition, rgr1 and sin4 mutants express promoters lacking UAS regions (22–24). UAS-independent promoter expression is also observed in histone H3 mutants or upon acute depletion of histone H4 (25, 26). Thus rgr1 and sin4 mutants may suffer an altered distribution or overall reduction of nucleosome density, which may lead to defects in transcriptional regulation. A second set of observations suggests that Rgr1p and Sin4p affect RNA polymerase II interaction with transcriptional activators more directly. Rgr1p and Sin4p are subunits of the Mediator (27), a protein complex associated with RNA polymerase II that is required for stimulation of transcription by activators in vitro (28, 29). rgr1 and sin4 mutations block repression by Rme1p (21), but it is unclear which aspect of the mutants’ defects impairs repression. Therefore, we have used a direct assay of the effect of Rme1p on activator binding as a step toward elucidation of the mechanism of repression.
MATERIALS AND METHODS
Yeast Strains and Plasmids.
Yeast strains have genetic markers ura3 leu2 trp1 lys2 ho::LYS2 and are isogenic except as noted. Culture conditions, media, strain constructions, transformations, and β-galactosidase assays followed standard recipes and protocols (13, 30). Rme1p was expressed from the GAL1 promoter (31) in functional form (PGAL1-S53-RME1 allele) or nonfunctional form (PGAL1-S53-rme1–213 allele). A gal80::LEU2 mutation permitted GAL1 promoter activity in the absence of galactose.
The CYC1-lacZ plasmids pAC153–4 and pAC142–10 have been described (13). Plasmid pAC153ΔRS was constructed from a CYC1-lacZ plasmid, pLGΔ312ΔRS, from which the TATA and RNA initiation sites had been deleted (32) through in vivo recombination (33). A fragment containing contiguous URA3 sequences, the RC, and the CYC1 UAS region was released from plasmid pAC153–4 through digestion with SphI and StuI, and then cotransformed into a ura3 mutant yeast strain with plasmid pAC153–4 that had been cleaved between URA3 and CYC1 UAS sequences with SmaI. Ura+ transformants were used to retrieve plasmid DNA in Escherichia coli, and restriction digestion confirmed that the retrieved plasmid had the predicted structure.
In Vivo Footprinting.
In vivo UV photofootprinting and dimethyl sulfate (DMS) footprinting were performed as described (34, 35). One-liter yeast cultures were grown in YPAc medium (1% yeast extract/2% bacto-peptone/2% potassium acetate) to early exponential phase, collected, and resuspended in 15 ml of fresh YPAc medium. For UV photofootprinting, 1.5-ml portions of the cell suspension were irradiated at 254 nm with a UV-crosslinker (Stratagene or Funakoshi, Tokyo), using doses of 500 or 1000 mJ/cm2, and DNA was isolated. As a control, purified DNA was irradiated using doses of 120 mJ/cm2. For DMS footprinting, 1.5-ml portions of the cell suspension were treated with 0.12% and 0.06% DMS for 2 min at room temperature, and DNA was isolated. As a control, purified DNA was methylated by 0.1% DMS for 3 min. The sites of UV photoproducts and methylation were analyzed by primer extension with primers RC1 (5′-TAGTTTAAAGAATTTGAACTATTTTTTGGCCGGTACC-3′, corresponding to −350 to −314 of plasmid pAC153–4), RC2 (5′TATGCCTGTATGTGTCAGCACTAAAGTTGCCTGG-3′, corresponding to −116 to −164 of plasmid pAC153–4), and RC3 (5′-GACGTGGGTAGGAAAAAAGTGAGCGCCAACACGGTACC-3′, corresponding to −481 to −444 of plasmid pAC142–10). We have numbered CYC1 upstream nucleotides according to refs. 36 and 37, in which position −208 corresponds to position −205 of ref. 38.
RESULTS
To explore the mechanism of transcriptional repression by Rme1p, we analyzed DNA–protein interactions in a CYC1-lacZ plasmid carrying the RC (RC-CYC1-lacZ gene, pAC153–4; Fig. 1B). CYC1 expression depends on the activators Hap1p, a Zn2-Cys6 zinc cluster protein that binds to UAS1 at −269 to −247, and Hap2p/3p/5p, the yeast CCAAT-binding complex that binds to UAS2 at −210 to −203 (38, 39). We examined yeast strains in which RC-CYC1-lacZ was derepressed. We examined both RME1 strains, in which the low level of natural RME1 expression in growing cells is insufficient for repression (data not shown), and PGAL1-rme1–213 strains, which overexpress a nonfunctional Rme1p mutant protein (13, 31). Hap1p binding was detectable by in vivo UV photofootprinting, in which presence of a DNA-binding protein can either increase or decrease the formation of pyrimidine dimers upon irradiation of whole cells, compared with isolated DNA. Strong enhancements of UV photoproducts were observed at UAS1 nucleotides −257 and −258 in cells from a HAP1 wild-type strain (Fig. 2A, lanes 7 and 9) but not in cells from an isogenic hap1Δ deletion mutant (Fig. 2A, lanes 10 and 12) and not in naked DNA (Fig. 2A, lanes 8 and 11). The position and genetic requirements for these enhancements in UAS1 demonstrate that they depend on Hap1p binding in vivo.
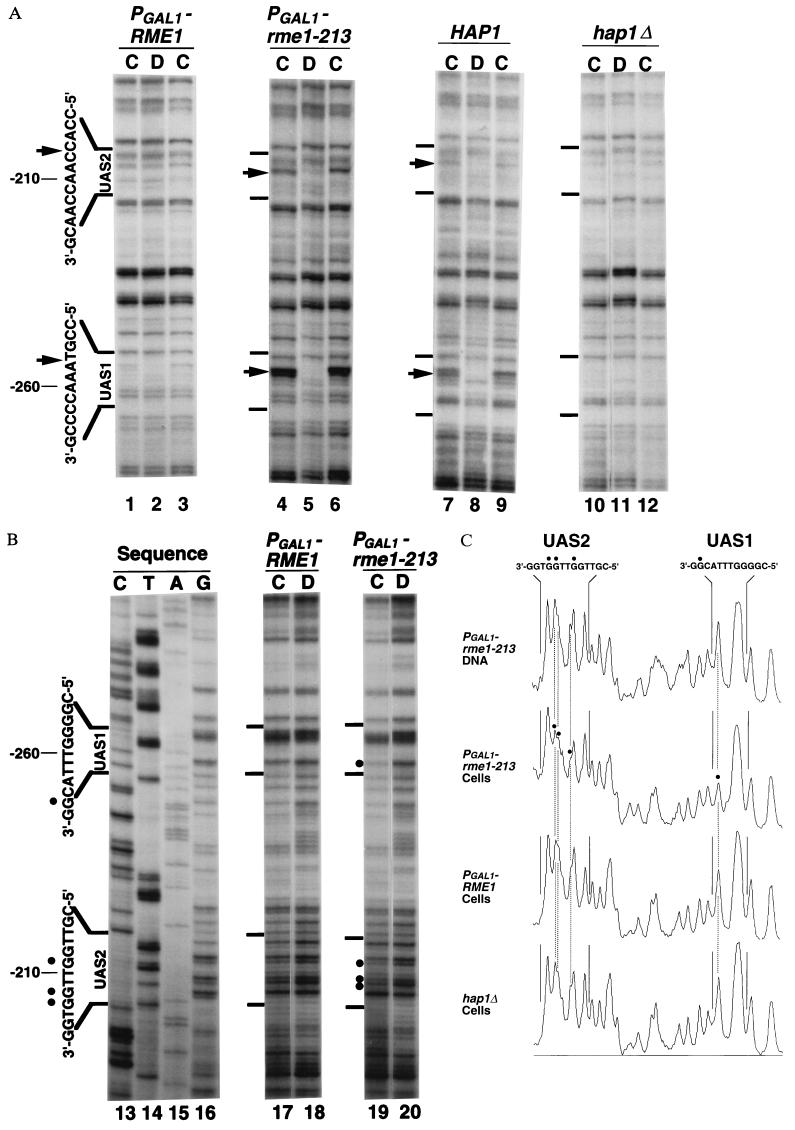
Figure 2.
(A) UV photofootprints of the noncoding strand of UAS1 and UAS2 regions in plasmid pAC153–4. UAS sequences are shown to the left of the gels. Arrows indicate sites of enhancements of UV photoproducts in irradiated cells (lanes C) compared with irradiated purified DNA (lanes D). Lanes 1–3, strain AMP1122 (α rme1::PGAL1-S53-RME1::TRP1 gal80::LEU2); lanes 4–6, strain AMP1124 (α rme1::PGAL1-S53-rme1–213::TRP1 gal80::LEU2); lanes 7–9, strain AMP108 (α); lanes 10–12, AMP1434 (α hap1Δ::LEU2). All strains have additional markers ura3 leu2 trp1 lys2 ho::LYS2 and carry plasmid pAC153–4. (B) In vivo DMS footprints of coding strand of UAS1 and UAS2 in plasmid pAC153–4. DMS-treated DNA (lanes 17 and 18, strain AMP 1122; lanes 19 and 20, strain AMP 1124) was subjected to primer extension mapping. Lanes 17 and 19 are samples from DMS-treated intact cells; lanes 18 and 20 are samples of DNA purified before DMS treatment; lanes 13–16 are dideoxy sequencing samples. Sequencing reactrions were terminated with the complementary dideoxynucleotide; for example, lane 16 (G) was terminated with dideoxy-CTP. The UASs are shown to the left of the gels. Dots indicate sites protected from DMS modification in cells. (C) Densitometric scans of DMS footprints shown in B: lane 20 (PGAL1-rme1–213 DNA), lane 19 (PGAL1-rme1–213 cells), lane 17 (PGAL1-RME1 cells), and a lane on the same gel containing a sample of DMS-treated AMP1434 cells (hap1Δ cells).
Hap1p binding was also detected through in vivo DMS footprinting, in which binding of a protein to DNA blocks methylation of G residues. We observed protection of the G residue at nucleotide −255 in UAS1 [Fig. 2B, lane 19 (cells) compared with lane 20 (purified DNA)]. This base was protected from DMS modification in vitro by purified Hap1p (40). Protection of this G residue was relieved by a hap1Δ mutation (Fig. 2C, hap1Δ cells compared with PGAL1-rme1–213 cells). Thus, DMS protection at −255 results from Hap1p binding in vivo.
We determined whether Rme1p could inhibit DNA binding by Hap1p through in vivo footprinting of plasmid pAC153–4 in PGAL1-RME1 strains, in which RC-CYC1-lacZ expression is repressed by functional Rme1p (Fig. 1A). Presence of functional Rme1p caused a severe reduction in binding of Hap1p to UAS1, on the basis of both UV photofootprinting (Fig. 2A, lanes 1 and 3) and DMS footprinting assays [Fig. 2 B, lane 17 (cells) compared with lane 18 (purified DNA), and C, PGAL1-RME1 cells compared with PGAL1-rme1–213 cells].
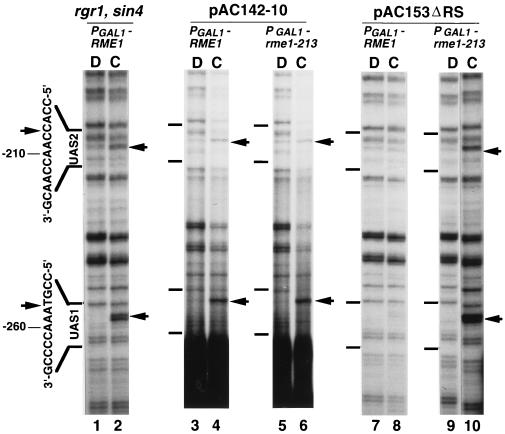
To determine whether Hap1p binding inhibition was relevant to the mechanism of repression by Rme1p, we examined effects of two types of mutations that relieve repression but do not affect binding of Rme1p to DNA. First, a deletion of RC sequences (−443 to −313, corresponding to IME1 sequences −1873 to −1743) abolishes repression (Fig. 1B, plasmid pAC142–10). This deletion does not remove either Rme1p binding site. We observed the same level of Hap1p binding to plasmid pAC142–10 in both PGAL1-RME1 and PGAL1-rme1–213 strains, on the basis of UV photofootprinting [Fig. 3, lanes 4 and 6 (cells) compared with lanes 3 and 5 (purified DNA)]. Second, rgr1 and sin4 mutations cause a defect in Rme1p-dependent repression, and we have since found that an rgr1 sin4 double mutant has a similar repression defect (M.S., W.L., P. A. Covitz, H.S., and A.P.M., unpublished work). In vivo footprinting studies indicate that binding of Rme1p to the two sites in the RC is unaffected by the rgr1 sin4 defect (M.S., W.L., P. A. Covitz, H.S., and A.P.M., unpublished work). We observed efficient Hap1p binding to UAS1 in a PGAL1-RME1 rgr1 sin4 strain, on the basis of UV photofootprinting [Fig. 3, lane 2 (cells) compared with lane 1 (purified DNA)]. Therefore, inhibition of Hap1p binding by Rme1p and transcriptional repression by Rme1p share the same genetic requirements.
Figure 3.
Effects of cis- and trans-acting mutations that disrupt repression by Rme1p on binding of Hap1p and Hap2p/3p/5p. In vivo UV photofootprinting of the noncoding strand was performed with intact cells (lanes C) or with purified DNA (lanes D) as in Fig. 2. UASs are shown to the left of the gels. Arrows indicate sites of enhancements of UV photoproducts in cells. Lanes 1 and 2, strain AMP1426 (α rme1::PGAL1-S53-RME1::TRP1 gal80::LEU2 rgr1–100 sin4::TRP1) carrying plasmid pAC153–4; lanes 3 and 4, strain AMP1122 carrying plasmid pAC142–10; lanes 5 and 6, strain AMP1124 carrying plasmid pAC142–10; lanes 7 and 8, strain AMP1122 carrying plasmid pAC153ΔRS; lanes 9 and 10, AMP1124 carrying plasmid pAC153ΔRS.
We also attempted to examine binding of Hap2p/3p/5p to UAS2 in these experiments. Enhancement of UV photoproducts was detectable at UAS2 nucleotides −207 and −208 in PGAL1-rme1–213 cells (Fig. 2A, lanes 4 and 6) but not in purified DNA (Fig. 2A, lane 5). We also observed weak protection from DMS of G residues at −211, −208, and −207 in UAS2 [Fig. 2B, lane 19 (cells) compared with lane 20 (purified DNA)]. No studies of DMS protection by Hap2p/3p/5p have been reported, but methylation at −208 and −207 interferes with Hap2p/3p/5p binding in vitro (38, 39). Both UV and DMS footprinting assays suggest that binding to UAS2 is reduced in PGAL1-RME1 cells (Fig. 2). However, we were unable to verify that a hap2Δ mutation eliminates these footprints because the strain was unable to grow in the medium used for this analysis (YPAc); thus, we cannot be certain that these footprints represent Hap2p/3p/5p binding.
Our observations argue that Rme1p, in conjunction with Rgr1p and Sin4p, exerts repression through inhibition of binding of transcriptional activators. Rgr1p and Sin4p have recently been identified as subunits of the RNA polymerase II-associated Mediator complex (27). We envisioned that Rgr1p and Sin4p might serve as a bridge between Rme1p and RNA polymerase II; the resulting complex might then be tethered both upstream of UASs (at the Rme1p sites) and downstream of UASs (through basal transcription machinery at the TATA and RNA initiation region). This hypothetical complex might exert steric constraints on the UAS region that block activator binding indirectly, as a consequence of a repressive Rme1p–RNA polymerase II interaction. To evaluate this possibility, we analyzed an RC-CYC1-lacZ-derived plasmid that includes the RC, UAS1, and UAS2, but lacks the TATA and RNA initiation sites in the CYC1 promoter (Fig. 1B, plasmid pAC153ΔRS, with a deletion of −183 to +40). Absence of essential promoter sequences prevents RC-CYC1-lacZ expression from this plasmid. UV photofootprinting in PGAL1-RME1 and PGAL1-rme1–213 strains indicated that Rme1p inhibits Hap1p and Hap2p/3p/5p binding to UAS1 and UAS2 in this plasmid [Fig. 3, lanes 8 and 10 (cells) compared with lanes 7 and 9 (purified DNA)]. Thus activator exclusion by Rme1p does not require known basal promoter sequences at CYC1.
DISCUSSION
We have provided direct evidence that Rme1p can act over a 250- to 300-bp distance to prevent activators from binding to target sites in vivo, as summarized in a model for repression of the RC-CYC1-lacZ hybrid gene by Rme1p (Fig. 4). In the absence of Rme1p, the activator Hap1p is bound to its target site, UAS1. In the presence of Rme1p, Rgr1p-Sin4p, and intact RC sequences, Hap1p is not bound to UAS1. We refer to this repression mechanism as activator exclusion. We argue that activator exclusion is the mechanism by which Rme1p exerts repression because of the correlation between their genetic requirements. Both activator exclusion and repression depend upon Rgr1p and Sin4p and upon RC sequences that flank the Rme1p binding sites. Both indirect (21) and direct (M.S., W.L., P. A. Covitz, H.S., and A.P.M., unpublished work) experiments indicate that Rme1p is bound to its sites in rgr1 and sin4 mutants and in the absence of the −1873/−1734 interval. Thus the requirements for activator exclusion are not trivial requirements for Rme1p expression or DNA binding ability.
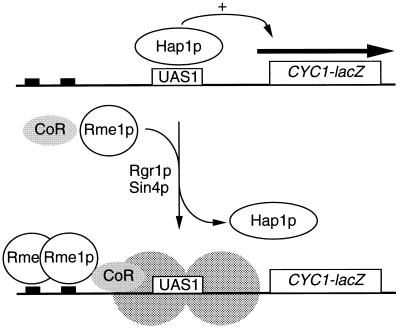
Figure 4.
Model for repression by Rme1p. In the absence of Rme1p, the activator Hap1p can bind to UAS1 and activate transcription (Upper). When Rme1p is present, it binds to upstream sites and alters the template to prevent activator binding (Lower). Repression and activator exclusion depend upon sequences flanking Rme1p binding sites, which may include binding sites for a hypothetical corepressor (designated CoR). Activator exclusion also depends upon Rgr1p and Sin4p, and may result from an altered state of chromatin (depicted by shaded circles).
Rme1p may inhibit activator binding through an effect on chromatin structure. Rgr1p and Sin4p are required to maintain high nucleosome density (22, 23), which may be required for Rme1p to establish a repressive chromatin structure. Rme1p apparently acts through a different mechanism than α2-Mcm1p, the repressor of yeast a-specific genes (reviewed in refs. 2 and 5); repression by α2-Mcm1p is only slightly dependent upon Rgr1p and Sin4p (24, 41), whereas repression by Rme1p is almost completely dependent upon Rgr1p and Sin4p (21). In vitro experiments indicate that α2-Mcm1p may repress through effects on the basal transcription machinery (42), and α2-Mcm1p does not prevent the activator Gal4p from binding to a nearby site in vivo (43). There is also a clear correlation between repression by α2-Mcm1p and local positioning of nucleosomes (34, 35, 44). Histone function and interaction with the α2-Mcm1p corepressor, Tup1p, are required for full repression by α2-Mcm1p (5, 34, 45), but it is unclear thus far what the biochemical consequences of an α2-Mcm1p-directed chromatin change may be. Our studies of Rme1p indicate that activator exclusion is the biochemical consequence of repression, but it is uncertain that Rme1p causes a chromatin structural change.
It is uncertain why RC sequences flanking the Rme1p binding sites are required for activator exclusion. One simple explanation is that Rme1p acts together with a DNA-bound corepressor in establishing repression, as illustrated in Fig. 4. A second explanation is that a length of free DNA is required for Rme1p to establish a stable, repressive structure. We favor the first possibility because point mutations outside of the Rme1p binding sites can impair repression (W.L., P. A. Covitz, and A.P.M., unpublished observations). However, we have not identified the hypothetical corepressor.
Activator exclusion resembles transcriptional silencing at the yeast HM loci and at telomeres (reviewed in refs. 4 and 46). However, we believe that these mechanisms are distinct because the IME1 locus is far from telomeric sequences and because Rme1p repression is unaffected by sir2, sir3, or sir4 null mutations (14), which abolish telomeric and HM locus silencing. Rme1p may repress through a similar mechanism to Drosophila Polycomb-group (Pc-G) gene products, which maintain repression of homeotic genes in inappropriate cell types (47, 48). Repression elements that respond to Pc-G products are orientation-dependent (47), as is the RC (14). In addition, polytene chromosome immunocytochemistry and activation assays indicate that Pc-G repression can inhibit binding of the trans-activator Gal4p to chromosomal target sites (47, 48). Thus repression by Pc-G products and by Rme1p cause activator exclusion.
It remains difficult to reconcile the roles of Rgr1p-Sin4p in Mediator function, chromatin structure, and Rme1p repression. We note that a chromatin remodeling complex, the SWI-SNF complex (49), cofractionates with RNA polymerase II (50). The purified SWI-SNF complex can alter a chromatin template to improve interaction between DNA and DNA binding proteins (51, 52). Thus it is possible that the Mediator can alter chromatin structure through an effect on SWI-SNF activity. A second possibility is that Rgr1p and Sin4p affect chromatin structure indirectly, for example, through an effect on expression of chromatin structural components. Identification of additional mutations that disrupt repression by Rme1p may help to resolve these models.
Acknowledgments
We thank the members of the Mitchell laboratory for helpful advice and discussion, and Drs. U. Matsumoto and Y. Shibusawa for their support in the course of this work. We thank our colleagues T. Bestor, M. Carlson, D. Shore, and S. J. Silverstein for comments on this manuscript. This work was funded by Grants-in-Aid for Scientific Research on Priority Areas and for Encouragement of Young Scientists from the Ministry of Education, Science and Culture, Japan and by a Short-Term Fellowship from the Human Frontier Science Program Organization (SF-266/93) to M.S., and by Public Health Service Grant GM39531 and an American Cancer Society Faculty Research Award to A.P.M.
Footnotes
Abbreviations: UAS, upstream activation sequence; DMS, dimethyl sulfate; RC, Repression Cassette.
References
- 1.Johnson A D. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 2.Herschbach B M, Johnson A D. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 3.Hanna-Rose W, Hansen U. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 4.Laurenson P, Rine J. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth S Y. Curr Opin Genet Dev. 1995;5:168–173. doi: 10.1016/0959-437x(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 6.Kassir Y, Simchen G. Genetics. 1976;82:187–206. doi: 10.1093/genetics/82.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rine J D, Sprague G G, Jr, Herskowitz I. Mol Cell Biol. 1981;1:958–960. doi: 10.1128/mcb.1.10.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honigberg S M, McCarroll R M, Esposito R E. Curr Opin Cell Biol. 1993;5:219–225. doi: 10.1016/0955-0674(93)90106-z. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell A P. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell A P, Herskowitz I. Nature (London) 1986;319:738–742. doi: 10.1038/319738a0. [DOI] [PubMed] [Google Scholar]
- 11.Kassir Y, Granot D, Simchen G. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 12.Smith H E, Mitchell A P. Mol Cell Biol. 1989;9:2142–2152. doi: 10.1128/mcb.9.5.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covitz P A, Mitchell A P. Genes Dev. 1993;7:1598–1608. doi: 10.1101/gad.7.8.1598. [DOI] [PubMed] [Google Scholar]
- 14.Covitz P A. Ph.D. thesis. New York: Columbia University; 1993. [Google Scholar]
- 15.Toone W M, Johnson A L, Banks G R, Toyn J H, Stuart D, Wittenberg C, Johnston L H. EMBO J. 1995;14:5824–5832. doi: 10.1002/j.1460-2075.1995.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith H E, Su S S Y, Neigeborn L, Driscoll S E, Mitchell A P. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman A, Shefer M, Sagee S, Kassir Y. Mol Gen Genet. 1993;237:375–384. doi: 10.1007/BF00279441. [DOI] [PubMed] [Google Scholar]
- 18.Zitomer R S, Lowry C V. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson A D. In: A Combinatorial Regulatory Circuit in Budding Yeast. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 975–1006. [Google Scholar]
- 20.Johnston M, Carlson M. In: Regulation of Phosphate and Carbon Utilization. Jones E W, Pringle J R, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 21.Covitz P A, Song W, Mitchell A P. Genetics. 1994;138:577–586. doi: 10.1093/genetics/138.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y W, Dohrmann P R, Stillman D J. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y W, Stillman D J. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, West R W, Jr, Johnson S J, Gans H, Kruger B, Ma J. Mol Cell Biol. 1993;13:831–840. doi: 10.1128/mcb.13.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han M, Grunstein M. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 26.Prelich G, Winston F. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Bjorklund S, Jiang Y W, Kim Y-J, Lane W S, Stillman D J, Kornberg R D. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 29.Bjorklund S, Kim Y-J. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 31.Covitz P A, Herskowitz I, Mitchell A P. Genes Dev. 1991;5:1982–1989. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- 32.Bowdish K S, Mitchell A P. Mol Cell Biol. 1993;13:2172–2181. doi: 10.1128/mcb.13.4.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma H, Kunes S, Schatz P J, Botstein D. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 34.Roth S Y, Shimizu M, Johnson L, Grunstein M, Simpson R T. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 35.Simpson R T, Roth S Y, Morse R H, Patterton H G, Cooper J P, Murphy M, Kladde M P, Shimizu M. Cold Spring Harbor Symp Quant Biol. 1993;58:237–245. doi: 10.1101/sqb.1993.058.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Guarente L, Mason T. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- 37.Pfeifer K, Prezant T, Guarente L. Cell. 1987;49:19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- 38.Olesen J, Hahn S, Guarente L. Cell. 1987;51:953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- 39.McNabb D S, Xing Y, Guarente L. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Guarente L. Genes Dev. 1994;8:2110–2119. doi: 10.1101/gad.8.17.2110. [DOI] [PubMed] [Google Scholar]
- 41.Wahi M, Johnson A D. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herschbach B M, Arnaud M B, Johnson A D. Nature (London) 1994;370:309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- 43.Redd M J, Stark M R, Johnson A D. Mol Cell Biol. 1996;16:2865–2869. doi: 10.1128/mcb.16.6.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu M, Roth S Y, Szent-Gyorgyi C, Simpson R T. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edmondson D G, Smith M M, Roth S Y. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 46.Shore D. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 139–191. [Google Scholar]
- 47.Zink D, Paro R. EMBO J. 1995;14:5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCall K, Bender W. EMBO J. 1996;15:569–580. [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson C L, Tamkun J W. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 50.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 51.Cote J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 52.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]