Abstract
Objective:
To determine whether specific genetic variations in the mtDNA that impact energy production and free-radical generation are potential new risk factors for in-hospital mortality after severe trauma.
Summary Background Data:
Each of the 3 mitochondrial DNA polymorphisms selected for this study (at positions 4216, 10398, 4917) alter the amino acid sequence of different key subunits of Complex I in the electron transport chain. They have been previously implicated in phenotypes involving tissues with high-energy demand, such as the brain and retina.
Methods:
Seven hundred forty-five consecutive patients admitted to the trauma intensive care unit at Vanderbilt University Medical Center between April 11, 2005, and February 27, 2006, were potentially eligible for this study. Under an Institutional Review Board-approved protocol (which excluded patients <18 years of age and prisoners), 666 patients had DNA extracted from a blood sample. Detailed demographic and clinical covariates were also obtained (including age, gender, ethnicity, lactate measurements, and injury severity score). A flurogenic 5′ nuclease allelic discrimination Taqman assay and the ABI 7900HT Sequence Detection System (v2.1) was used to genotype the T4216C, A10398G, and A4917G polymorphisms. The primary outcome was in-hospital mortality.
Results:
Multivariate logistic regression analysis revealed that the 4216T allele was a significant independent predictor of in-hospital mortality (OR = 2.63, 95% CI 1.14–6.07, P = 0.02) after adjustment for age, gender, injury severity score, highest lactate level, mechanism of injury, and the 10398 polymorphism.
Conclusions:
Variation in the mtDNA, specifically the 4216T allele, appears to increase the risk of in-hospital mortality after severe injury.
Humans have 2 genomes, the large nuclear genome and a smaller genome residing in the mitochondria. Genes encoded in the mitochondrial DNA are vital for energy production. In this study, a specific polymorphic variation in the mitochondrial genome (mtDNA) (4216T) is associated with in-hospital mortality after admission to a trauma intensive care unit.
The stress response after injury is characterized by high-energy requirements and inflammation. About 90% of all cellular adenosine 5′-triphosphate is produced by the electron transport chain embedded in the inner mitochondrial membrane.1–3 This chain of multisubunit protein complexes consist of gene products from the 2 human genomes: the mitochondrial and nuclear.4 The mitochondrial genome (mtDNA) consists of only 16,569 base pairs, yet it encodes 13 critically important subunits of the electron transport chain.1,4,5
Mitochondria are a major source of oxygen-derived free radicals, also collectively known as reactive oxygen species (ROS) and therefore, contribute to the inflammatory response. Under normal physiologic conditions, as many as 2% of electrons leak form the mitochondrial electron transport chain and reduce oxygen to superoxide anion, triggering formation of a cascade of free radicals that indiscriminately damage biologic macromolecules.3,6–10 Mitochondria are especially susceptible to damage by ROS because the mtDNA has limited reparative capacity.1 Damage to mtDNA creates a cycle of worsening mitochondrial dysfunction and increased ROS production. Both cellular energy production and damage by ROS have long been thought to play roles in the stress response and the resulting patterns of secondary injury.
Stable single nucleotide polymorphisms (SNPs) have emerged in mtDNA over the past 150,000 years.11 The distribution of these mtDNA polymorphisms vary greatly across populations reflecting human migration and adaptation to environmental conditions.12–16 The result is that in humans there are different types of mitochondrial electron transport chains defined by SNPs that alter the amino acid sequences of critical subunits in these protein complexes. Each of these types may have slightly different capacities for energy production and free-radical generation. Variations in mitochondrial SNPs have been associated with Parkinson disease, Alzheimer disease, Friedrich’s ataxia, amyotrophic lateral sclerosis.16–22 In this study we selected 3 specific SNPs located at positions 4216, 10398, and 4917 in the mtDNA because they alter the amino acid sequence of different key subunits of Complex I and because they have been implicated in other phenotypes involving high-energy demand.18,19,23–32
We hypothesized that specific mitochondrial genetic polymorphisms that alter electron transport could lead to impaired energy production or increased ROS production resulting in increased mortality after traumatic injury. To test this hypothesis we studied a large, consecutive cohort of trauma patients enrolled in the Vanderbilt Trauma Genetics Repository.
METHODS
Study Population
All patients admitted the Vanderbilt University Medical Center (VUMC) trauma intensive care unit (ICU) from April 11, 2005 to February 27, 2006, were potentially eligible for this prospective study cohort. VUMC is located in Nashville, Tennessee, and is the only level I trauma center serving an 80,000 square-mile catchment area. Individuals younger than 18 years old, prisoners, those with known pregnancy, and those that expired before admission to the Trauma ICU were excluded. All patients had a blood sample drawn for DNA extraction within 24 hours of admission to the Trauma ICU. The primary outcome for this particular study was in-hospital mortality, specifically death occurring before discharge from this hospital after admission to the VUMC Trauma ICU. All patients were enrolled in this study under a protocol approved by the VUMC Institutional Review Board.
Data Sources
VUMC’s clinical and research information infrastructure provided the linked demographic, clinical, laboratory, genetic, and outcome data used in this study. This infrastructure has been described in detail elsewhere.33–35 Specific components used in this study include:
TRACS (Trauma Registry of the American College of Surgeons): All patients evaluated at VUMC for trauma or burns had their data entered into the TRACS database. Currently more than 300 parameters are captured via retrospective chart review. This study made use of patient demographic, clinical, laboratory, and outcome data from this registry.
Vanderbilt Trauma Genetic Repository: Since April 11, 2005, all consecutive Trauma ICU admissions have DNA samples and genotype data stored in this repository.
Covariates
Demographics
Patient age in years, gender, and primary mechanism of injury (blunt or penetrating) were obtained from TRACS. Ethnicity was categorized as white, African American, Hispanic, Asian, and other.
Physiologic Reserve
ISS and patient age, obtained from TRACS, were used to assess magnitude of injury and preinjury physiologic reserve. Laboratory measures of physiologic reserve used in this study included acidosis and hemorrhage severity, as reflected in the admission hematocrit. Acidosis was assessed using 3 measurements: initial lactate value, highest lactate value in the first 24 hours, and the highest lactate over the entire hospitalization.
DNA Analysis
DNA was isolated from whole blood using PUREGENE (Gentra Systems Inc., Minneapolis, MN). Genotyping was performed with the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems Inc., Foster City, CA) using the 5′nuclease allelic discrimination Taqman assay. The mtDNA 4216C SNP was detected using the ABI sequence detection system. Primer and probe sequences are as follows: TaqMan MGB probe for the T allele: 6-FAM-AGC ATT ACT TAT ATG ATA TGT C; TaqMan MGB Probe for the C allele: VIC-TAG CAT TAC TTA TAT GAC ATG TC; 4216 Forward Primer, TCC TAT GAA AAA ACT TCC TAC CAC TCA; 4216 Reverse Primer GCT GGA GAT TGT AAT GGG TAT GG. The mtDNA 10398A SNP was also detected using the ABI sequence detection system. Primer and probe sequences are as follows: 10398 Forward Primer: CTA CAA ACA ACT AAC CTG CCA CTA ATA G; 10398 Reverse Primer: GGG CAT TTG GTA AAT ATG ATT ATC A; Taqman MGB Probe for G allele: VIC-AGA CTG AGC CGA ATT; Taqman MGB Probe for A allele: 6FAM-TAG ACT GAA CCG AAT TG. The mtDNA 4917G SNP was detected using a MGB Eclipse Probe because of a polymorphism at the position 4918 (Nanogen, Bothell, WA). The 4917 forward primer was CAACTGCCTGCTA*TGATGGAT and the 4917 reverse primer was GGCCTGCTTCTTCTCACATGACA (* represents a proprietary nucleotide analog).The FAM probe for the wild type 4917 allele was TTACGA*TT*A*GT*GN*GG and the TET probe for the 4816G allele was TTACGA*CTA*GT*GN*GG (with N and * representing proprietary nucleotide analogs.) Genotypic data were analyzed using ABI Sequence Detection System version 2.1 software and confirmed by visual inspection of the plots. Genotypes were classified as undetermined if PCR amplification failed with the specified sets of probes and primers.
Statistical Analysis
Mitochondrial genotype frequencies and other categorical variables were compared between survivors and nonsurvivors using the Pearson χ2 test or the Fisher exact test. Continuous demographic variables were compared using the Student t test or the Mann-Whitney U test if the variables were not normally distributed. The Pearson product-moment correlation was used to measure the strength of association between 2 continuous variables. The mitochondrial polymorphisms evaluated in this study are generally homoplasmic (only 1 allele is present in a given individual). Unconditional multivariate logistic regression models were constructed to determine the odds ratios and 95% confidence levels for associations between the specific mitochondrial polymorphisms, potential confounding variables (such as injury severity score) and the outcome variable, in-hospital mortality. Tests for statistical significance were 2-sided with an α level of 0.05. All statistical analyses were accomplished using the STATA statistical software package (version 9.0; College Station, TX).
RESULTS
Study Population Characteristics
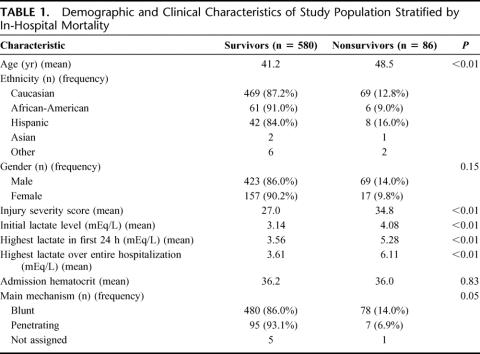
The initial cohort consisted of 745 patients. Seventy-nine patients met exclusion criteria. The remaining 666 patients comprise the study population. The demographic and clinical characteristics of this group are stratified by the primary outcome variable, in-hospital mortality, and presented in Table 1. The overall in-hospital mortality for this group was 12.9% (86/666). Age (mean, in years) was significantly different between survivors and nonsurvivors, 41.2 years versus 48.5, P < 0.001. Males represented 73.9% of this cohort (492/666). Although males had a higher rate of in-hospital mortality than females (14.0% vs. 9.8%), the difference was not statistically significant in this population (P = 0.15). ISS, initial lactate level, highest lactate level within 24 hours and highest lactate level over the entire hospitalization were all significantly elevated in nonsurvivors as compared with survivors. There was a modest, but statistically significant, correlation (r = 0.2, P < 0.001) between ISS and the highest lactate level over the entire hospitalization. In-hospital mortality also differed by primary mechanism of injury, with blunt trauma having a rate of 14.0% (78/558) compared with 6.9% (7/102) after penetrating trauma (P = 0.05). As expected, ISS varied significantly between the blunt trauma (mean = 29.5) and penetrating trauma (mean = 19.9; P < 0.01).
TABLE 1. Demographic and Clinical Characteristics of Study Population Stratified by In-Hospital Mortality
Mitochondrial Genotyping Results
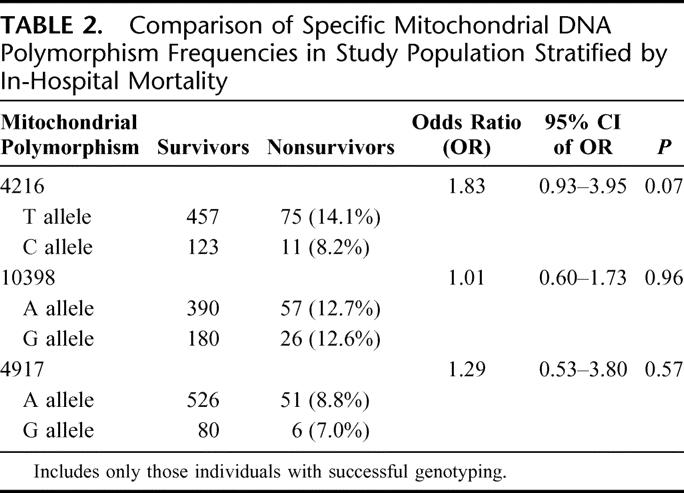
Genotyping efficiency for the 3 mitochondrial DNA polymorphisms in this study population was 4216 (99.0%), 10396 (98.1%), and 4917 (99.6%). Table 2 presents the results of the univariate analysis for each polymorphism. Of the 3 tested, the 4216 polymorphism revealed a possible association with in-hospital mortality. The T allele of 4216 had an in-hospital mortality rate of 14.1% (75/532) as compared with the 4216C allele, which had a mortality rate of 8.2% (11/134), OR = 1.83 (95% CI 0.93–3.95), P = 0.07.
TABLE 2. Comparison of Specific Mitochondrial DNA Polymorphism Frequencies in Study Population Stratified by In-Hospital Mortality
The distribution of these specific mitochondrial polymorphisms varied considerably by ethnicity. (Table 3) The C allele of 4216 was present in 22.9% (122/532) of white individuals, whereas it was found in only 4.5% (3/66) of African Americans and in 0% (0/50) Hispanics. The A allele of 10398 was present in 75.9% (403/531) of white individuals compared with 1.5% (1/65) of African Americans. The 4917 polymorphism also had a significantly varied distribution again based on ethnicity with the G allele being present in 10.6% (57/535) of whites and in none of the African Americans (0/67) or Hispanics (0/65). Because of the absence of polymorphic variation at these 3 mitochondrial sites in the African American and Hispanic groups, we confined the remainder of our analysis to white individuals. In whites (n = 532), the univariate analysis of the 4216 allele continued to reveal a possible association with in-hospital mortality. Those trauma victims with the 4216T allele had a mortality rate of 14.4% (59/410) and those with the C allele had a mortality rate of 8.2% (10/122), OR = 1.88 (95% CI 0.91–4.26), P = 0.07. The 4216T allele group had higher mean lactate levels than the 4216C allele group at time of admission (3.13 ± 1.9 vs. 3.02 ± 2.1), during the first 24 hours after admission (3.66 ± 2.5 vs. 3.39 ± −2.0), and during the entire hospitalization (3.80 ± 2.7 vs. 3.72 ± 2.7). However, these differences in lactate levels between genotype groups were not statistically significant in this study population.
TABLE 3. Ethnic Distribution of Specific Mitochondrial DNA Polymorphism Frequencies

There were also no statistically significant differences between the 4216T allele group compared with those with 4216C with regard to age (mean age: T allele = 43.7 years vs. C allele = 46.1, P = 0.21), gender (T allele = 74% male vs. C allele = 71% male, P = 0.38) or ISS (mean ISS score: T allele = 28.4 vs. C allele = 28.2, P = 0.82). A preliminary analysis of 2 secondary endpoints, ICU days, and total hospital days, reveals statistically significant differences based on 4216 genotype. The median number of ICU days for the 4216T allele group was 5.3 days compared with 6.1 day for the 4216C allele group (P = 0.04). A similar difference was also noted in median total hospital days [4216T allele group = 13.9 days vs. 4216C allele group = 15.2 days, (P = 0.01)]. We suspect that this difference is at least in part due to survival bias stemming from the statistically significant difference in-hospital mortality based on the 4216 genotype.
Multivariate Analysis
A multivariate logistic regression model was constructed to identify independent predictors of in-hospital mortality. Our primary model included age, gender, ISS, mechanism of injury, highest lactate level during hospitalization, and the 4216 and 10398 mitochondrial polymorphisms. The 4216 polymorphism was suspected as being a contributor to the outcome based on the univariate analysis. The 10398 polymorphism was also included in this model because the C allele of 4216 occurs with both of the 10398 alleles. Therefore, adjustment for the 10398 polymorphism by the model was necessary to discern if the 4216 polymorphism was truly an independent predictor of in-hospital mortality.
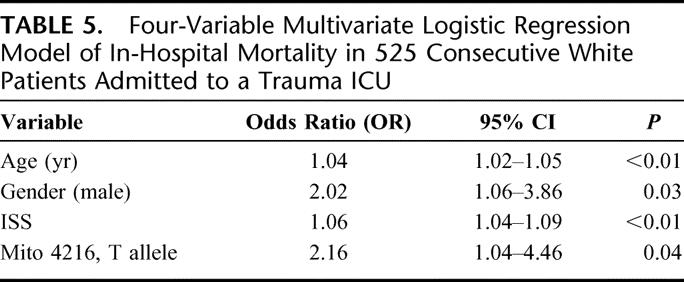
The results of this analysis are presented in Table 4. Not surprisingly, age (OR = 1.04, 95% CI 1.02–1.06, P < 0. 01), ISS (OR = 1.05, 95% CI 1.03–1.08, P < 0. 01) and highest lactate value during hospitalization (OR 1.25, 95% CI 1.14–1.35, P < 0. 01) are independent predictors of in-hospital mortality. This study also demonstrated that the mitochondrial DNA polymorphism 4216 was also an independent predictor (T allele: OR = 2.63, 95% CI 1.14–6.07, P = 0.02) of in-hospital mortality after adjusting for potential confounding variables (age, ISS, highest lactate level during hospitalization, plus gender, mechanism of injury, and the mitochondrial DNA polymorphism 10398). To confirm the robustness of this finding, a cut down model (Table 5) with just age, gender, and ISS as covariates demonstrated again that the mitochondrial DNA polymorphism 4216 remained an independent predictor of in-hospital mortality after severe trauma (T allele: OR = 2.16, 95% CI 1.04–4.46, P = 0.04).
TABLE 4. Multivariate Logistic Regression Model of In-Hospital Mortality in 525 Consecutive White Patients Admitted to a Trauma ICU
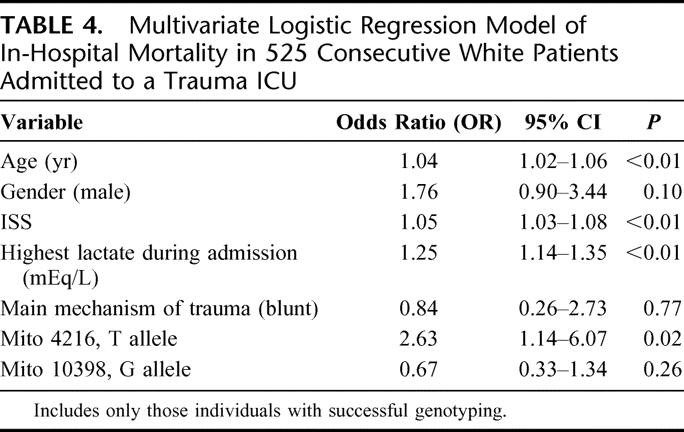
TABLE 5. Four-Variable Multivariate Logistic Regression Model of In-Hospital Mortality in 525 Consecutive White Patients Admitted to a Trauma ICU
DISCUSSION
After severe trauma, a very complex stress response occurs that involves high-energy demand and inflammation. The high mortality associated with this phenotype makes understanding this intricate process imperative. The complexity is daunting; however, a systematic uncovering of environmental and genetic factors in this patients population will hopefully further reduce in-hospital mortality after admission to a Trauma ICU. In this study, we demonstrated for the first time that a specific variation in the mtDNA appears to influence the ability to survive after trauma. The effect size for the 4216 polymorphism was unexpectedly large (T allele: OR = 2.63, 95% CI 1.14–6.07, P = 0.02) and remained an independent predictor even after careful adjustment for many known potential confounding variables. Perhaps these results should not be surprising given the central role mitochondria play in cellular energy production, apoptosis, and free-radical generation.1,4
The mtDNA is a rich source of genetic variation.1,4 The careful selection of the 3 mitochondrial SNPs in this study was based on 2 factors: (1) the mitochondrial polymorphism must result in the nonsynonymous change in an amino acid in an important subunit of Complex I in the electron transport chain, and (2) there must be prior epidemiologic evidence of association with disease phenotypes involving diminished energy production and/or increased free-radical generation. In the first polymorphism, T4216C, a histidine is substituted for a tyrosine in the ND1 subunit of Complex I in the mitochondrial electron transport chain. The 4216C allele has been associated with male infertility caused by asthenozoospermia, DIAMOAD, (a rare disorder comprised of diabetes insipidus, diabetes mellitus, optic atrophy, and deafness.) and Leber’s hereditary optic neuropathy.23–27 The second mtDNA polymorphism evaluated in our study, A10398G, alters the ND3 subunit of NADH dehydrogenase by changing a threonine to an alanine. This SNP has been associated with neurodegenerative phenotypes and longevity.18,19,28–30 The third mitochondrial polymorphism we incorporated into this study was A4917G. This SNP results in a change in the amino acid sequence from asparagine to aspartic acid in the ND2 subunit of Complex I. This SNP appears to be an independent predictor of peripheral neuropathy complicating antiretroviral therapy in HIV patients.32 In the case of each SNP, the proposed pathophysiologic mechanism includes an increased rate of electron leakage from Complex I resulting in increased ROS, which in turn contributes to both mitochondrial and nuclear DNA damage resulting in mitochondrial dysfunction.36,37 Alternatively, the adverse effects could relate directly to the less efficient functioning of Complex I or a reduced ability of individuals with these alleles to respond to an oxidative challenge.18
The specific mitochondrial polymorphism associated with increased risk for in-hospital mortality in the present study is located at position 4216 in the mtDNA and has 2 alleles (a T allele and a C allele). Analysis of our population reveals that trauma patients with the T allele carry an increased risk of in-hospital mortality relative to the C allele. The 4216T allele is present in approximately 80% of whites.25,38–41 The 4216C allele in this analysis would necessarily be viewed as protective relative to the 4216T allele. The 4216C allele has previously been associated with chronic neurodegenerative disorders such as Parkinson disease and Leber’s Hereditary Optic Neuropathy.23–27 A study by Ruiz-Pessini et al demonstrated that the 4216C allele was associated with male infertility in Spain.23 Natural selection should have eliminated this 4216C allele from the population because of its impact on reproductive fitness. However, the variation is found in 20% of whites. Our study shows for the first time that this allele appears to play a role in surviving trauma and perhaps account for the persistence of this variation within the white population. We suspect that the complete explanation for this finding will likely be far more complex. Our future research must be directed toward measuring the functional consequences of this specific mitochondrial DNA variation that has been associated with 3 very disparate phenotypes: trauma survival, male infertility, and specific types of neurodegeneration.
Genetic association studies are fraught with potential limitations that we expressly attempted to avoid in the present study.42 First, a population-based, prospectively collected cohort of consecutive patients to a Trauma ICU from a large catchment area was used in the analysis to minimize the effect of selection bias. Second, the dramatic variation in the distribution of the mtDNA polymorphisms in the different ethnic populations required that the results be stratified by specific ethnic group to avoid the confounding effect of unidentified population substructure. Third, mtDNA polymorphisms were selected that had prior epidemiologic evidence of association with phenotypes where alteration in cellular energy production and ROS production are important lending further biologic plausibility to their potential involvement. It remains possible that the 4216T allele is in linkage disequilibrium with a more important causative polymorphism. An important next step, after validation of this observation in another large trauma population, will be detailed sequencing of complete mtDNA s coupled with genotyping of important nuclear SNPs relevant for mitochondrial function. Epistatic interactions between individual loci in the mtDNA as well as nuclear-mitochondrial gene interactions may emerge.43 A recent study by Johnson et al of genetic expression profiles in patients developing sepsis after trauma showed that there is not just a great deal to be learned about the genetic component of the physiologic response to injury, but also that genetic information may be used to predict important clinical outcomes.44
CONCLUSION
In summary, this study provides novel evidence that polymorphic variation in the mtDNA contributes to in-hospital mortality after severe traumatic injury. The magnitude of the risk associated with the 4216T allele suggests that this polymorphism should be considered as an important new genetic risk factor. As we contemplate genome-wide association studies in trauma that will encompass the 6,000,000,000 base pairs in the human diploid genome it is worth recalling that humans have 2 genomes. Variation in the 16,569 base pairs of mtDNA should not be overlooked in our quest to uncover important genetic factors central to the stress response after severe injury.
Discussions
Dr. Timothy R. Billiar (Pittsburgh, Pennsylvania): Canter and colleagues show a significant association between in-house mortality and the presence of the 4216T allele in mitochondrial DNA in 660 trauma patients.
Underlying person-to-person variability and mitochondrial function could be a potential factor determining how well traumatic stress is tolerated by an individual.
This hypothesis is astutely derived from literature, some of it generated by Dr. Canter himself, demonstrating that mutations in genes for mitochondrial targeted proteins, proteins encoding genes involved in energetics, are associated with disease states. Among the most notable are defects in Complex I, also known as NADH ubiquinone oxidoreductase. This is a 42-protein complex that shuttles electrons to Coenzyme Q. Seven of these proteins derive from mitochondrial DNA. Mutations in mitochondrial DNA deserve special attention because stable mutations are easily propagated due to the lack of homologous recombination in the mitochondrial genome.
Known mitochondrial Complex I mutations are associated with inherited defects in high-energy consuming organs including the brain and heart. One of the most severe forms, fatal fetal lactic acidosis, demonstrates the consequences of severely altering Complex I function.
The Canter study implies that milder defects only manifest under severe stress states. It would be remarkable if a single nucleotide change resulting in a single amino acid substitution in 1 of 42 proteins could act as an independent predictor of mortality. If true, a new milestone in our understanding has been achieved.
We have understood for some time that elevated circulating lactic acid levels, and perhaps even more importantly sustained lactic acidosis, are associated with greater mortality following injury. Although this is not a uniform observation, most studies in traumatized or burned patients point to an association of high initial lactic acid levels and sustained levels in patients who die. This includes even a very recent study published in 2007 in burn patients by Cochran and coworkers where lactate at 12 and 48 hours served as a predictor for mortality independent of age, total body surface, burn, or gender.
Let’s connect the dots. Could the tendency to generate more lactate be a consequence of our mitochondrial phenotype? This raises several key questions that are left unanswered. I have 4 questions for you.
Number 1, what were the lactate levels in the T allele group compared to the C allele group regardless of survivorship?
Number 2, was there an association between the T allele in combination and another predisposing factor? An example might be male gender.
Third, you report only mortality. Do other more subtle endpoints correlate with the T allele? For example, ICU days, length of stay, infectious complications, or days on a ventilator.
Finally, there is a clear racial association with mitochondrial DNA polymorphisms. You also note differences in survival between whites and African Americans in your studies. Because mitochondrial DNA mutations clearly separate into groups based on race, how likely is it that the differences are due to other factors associated with race and that the separation by DNA polymorphisms only result in a racial identification?
Dr. Jeffrey A. Canter (Nashville, Tennessee): We have just begun to probe the data set for variations in association with the T allele and lactate levels. There are some very interesting racial differences in lactate levels. Again, this is a prospective cohort study. So we are not cherry-picking patients. African Americans have a lower lactate level at the time of admission by almost 1 milliequivalent compared to whites. We do not fully understand the reason for that.
With regard to the T allele and gender, we have not done that analysis yet. We know that gender was not an independent predictor once you took T into account. However, we have not done the fine detail work on looking at gender; I think that is a very interesting place to go.
We have begun to look at endpoints other than just mortality. This was really our first cut at this data. Dr. Morris is going to present some data at Shock Trauma in a little over a month looking at dividing mortality into 2 different groups, up to 72 hours and after 72 hours. This is certainly no surprise to surgeons, because you know that early trauma mortality in the hospital is different than late mortality. I can say that it appears that this mitochondrial variation emerges as a more important factor in the late group rather than the early. But we didn’t have enough statistical power to make that conclusion because we had to cut our mortality group in half.
Finally, I would like to talk a little bit about the racial strategies in mitochondrial disease. As opposed to a lot of other genetics, race is a very important factor. The mitochondrial genomes are significantly different based on race. This analysis is only stratified for whites because it just so happened that the 3 variations we looked at were not represented in either African Americans or Hispanics.
A lot of our work focuses on other racial groups. I have just published a paper in Cancer Research looking at invasive breast cancer in African American women, which we find associated with another mitochondrial polymorphism. I think we are at the very tip of the iceberg in approaching this. We are just now looking at race using nuclear markers in conjunction with mitochondrial markers.
Dr. Donald E. Fry (Chicago, Illinois): This reminds me of a paper written about 20 years ago by Sorensen et al (N Engl J Med 1988; 318:727–32), which identified that the odds ratio of dying from infection are greater if one of your parents died of infection at a premature age. This was the trigger for many people investigating genetic links in our immunity that would implicate immune deficiency as the cause. You raise the interesting idea as to whether the adaptive response of the cell may be defective and that similar insults might result in some individuals being more vulnerable than others. It certainly raises the question as to whether Sorensen needed to look at the gender of the parent as yet an additional association.
I found interesting, if I read your data correctly, that the T allele was expressed 4 times more commonly than the C allele. And I find it very interesting that Mother Nature preserved a gene that gives us increased vulnerability to dying from stress. So I think that must be rationalized somehow. Genotype can be very deceptive. There are 4 times as many proteins as there are genes in our nuclear genome. It would be interesting to know whether that relationship exists for mitochondrial genes.
But what is the phenotypic evidence in your patients to show that they really were having an energy crisis? Did the T allele patients demonstrate more evidence of liver failure? Did they experience a higher incidence or greater evidence of liver enzyme elevations? Did they have more frequent kidney failure? Did they demonstrate evidence to support the idea of a greater degree of organ insufficiency as a consequence either of cellular necrosis or of inappropriate apoptosis?
Finally, you didn’t identify the patients’ cause of death. Did they die of sepsis or of shock? What were the real causes? Obviously, that would be of some significance in trying to interpret the real meaning of this genetic marker.
Dr. Jeffrey A. Canter (Nashville, Tennessee): We are just in the process of breaking down the mortality into subsets and so I cannot respond specifically. But there is this temporal association that I mentioned previously.
As far as markers for energy deficiency, and particularly the possibility for free-radical injury, that is part of our ongoing work. It is difficult to look for markers. In the mitochondrial world we resort at the bench to transmitochondrial cybrid systems to make those measurements.
Your first question is excellent and very astute. This particular allele seems to be the more common allele particularly in whites. And that question was brought up by the exact opposite point.
The C allele gets all the press. It is associated with infertility in 1 study from Spain and with the development of neurodegenerative disorders. The key point here is reproductive fitness. The T allele perhaps is associated with greater fertility. You may be less likely to survive trauma, but you are more fertile. From the genes point of view, this is all that matters. Hence, this genetic variation dominates in the white population. The C allele would be the flip-side of this. With C you may be more likely to survive trauma, but less reproductively fit. In the Spanish study, men with the 4216C allele were more likely to be infertile due to decreased sperm motility compared to 4216T.
The bottom line is that we must go to the bench and look at this particular genetic change. I do not think this is anything more than 1 bar of many bars in the genetic barcode of the response to trauma. But I think it certainly warrants further investigation.
Dr. Raymond Pollak (Naperville, Illinois): Cytokine gene polymorphisms are associated with outcome in certain inflammatory disorders, including organ transplant rejection, trauma, and the like. Since you have the specimens frozen, have you looked at cytokine gene polymorphisms and compared these data to what you have currently?
Secondly, as a logistic concern, how did you obtain informed consent from these patients for something so sensitive?
Dr. Jeffrey A. Canter (Nashville, Tennessee): We have not looked at any cytokine polymorphisms yet. This is our very first genetic testing in this study population. It certainly won’t be our last.
As to your second question about informed consent, this study was done without informed consent. It was done through an IRB-approved protocol at Vanderbilt that took approximately 9 months to craft. This is done under waiver of consent. And we made a strong argument with the attorneys at Vanderbilt that you cannot study severe trauma and its consequences in a consent environment. So this data was obtained without consent, but with IRB-approved protocol.
Dr. Christopher C. Baker (Boston, Massachusetts): My question is fairly simple. If you cannot answer Dr. Fry’s question about mortality maybe you can answer this one. Since the battle for shock is won or lost in mitochondria in the early phase, can you tell us whether there was a difference between shock on presentation in your groups? Obviously, you used the highest level of lactate as a marker. Can you just talk about shock and whether that made a difference?
Dr. Jeffrey A. Canter (Nashville, Tennessee): I wish I could answer that question. I think we are just at the stage of breaking down the data into those groups. We had a sample size of over 700 to start with. But, trauma is such a complex phenotype that as you break it down, you are left with multiple studies with sample sizes of 100, and inadequate power to make comment about associations. We actually looked carefully at early in-hospital mortality associated with shock. And we will present that data at Shock Trauma. It is just not quite ready for me to talk about yet.
Dr. Basil A. Pruitt, Jr. (San Antonio, Texas): You mentioned that this was an expensive assay. Before we all start measuring this in our patients, let me ask: in your multivariate analysis, when you determine the variance accounted for by age, ISS, lactate, and all the other variables listed, how much residual variance is accounted for by the Mito 4216T? Is it a significant amount? Lastly, could you expand a little bit on your statement that this has implications for managing critically ill patients? What do you envision that we are going to do with that?
Dr. Jeffrey A. Canter (Nashville, Tennessee): It takes money obviously to extract DNA and to set up the assays.
As far as the actual component of risk that can be attributed to this genetic variation, it is a little bit misleading. If you look at the odds ratio and it is 2, that just goes from C allele to the T allele. But if you look at ISS, it is 1.05. But that is 5% compounded for each point of ISS. I suspect that the risk associated with this single genetic variation is rather small compared to age and ISS. What we are going to do is prospectively look and test our model. This will answer the question.
Your other question asks about the implication of this finding. I am not a surgeon, but here is the point that I see. This is the same type of mitochondrial change that we see in retinal disease. And if you go to an ophthalmology meeting, we are talking about aggressive use of free-radical scavenging medications based on genotype. Another implication may relate to the administration of oxygen. If someone is genetically predisposed to form higher levels of oxygen-derived free radicals, then prolonged, high oxygen levels may not be a good thing. There are many other factors. But basically we are at the very first inning of the ball game, and I can only imagine management benefits from this finding.
Footnotes
Supported by grant RO1 HD047447-01 NICHD (JHM)
Reprints: Jeffrey A. Canter, MD MPH, Center for Human Genetics Research, 519 Light Hall, Vanderbilt University Medical Center, Nashville, TN 37212. E-mail: jeff.canter@vanderbilt.edu.
REFERENCES
- 1.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. Mitochondrial DNA in aging and disease. Sci Am. 1997;227:40–47. [DOI] [PubMed] [Google Scholar]
- 3.Papa S. Mitochondrial oxidative phosphorylation changes in the life span. Molecular aspects and physiopathological implications. Biochim Biophys Acta. 1996;1276:87–105. [DOI] [PubMed] [Google Scholar]
- 4.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. [DOI] [PubMed] [Google Scholar]
- 5.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. [DOI] [PubMed] [Google Scholar]
- 6.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci USA. 1988;85:6465–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halliwell B, Gutteridge JMC, eds. Free Radicals in Biology and Medicine. 3rd ed. Oxford: Oxford University Press; 1999. [Google Scholar]
- 9.Kang D, Hamasaki N. Mitochondrial oxidative stress and mitochondrial DNA. Clin Chem Lab Med. 2003;41:1281–1288. [DOI] [PubMed] [Google Scholar]
- 10.Scheffler IE. Mitochondria. New York: Wiley-Liss; 1999. [Google Scholar]
- 11.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. [DOI] [PubMed] [Google Scholar]
- 12.Torroni A, Huoponen K, Francalacci P, et al. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci USA. 1994;91:8739–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishmar D, Ruiz-Pesini E, Golik P, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. Epub December 30, 2002. [DOI] [PMC free article] [PubMed]
- 15.Chen YS, Torroni A, Excoffier L, et al. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet. 1995;57:133. [PMC free article] [PubMed] [Google Scholar]
- 16.Autere J, Moilanen JS, Finnila S, et al. Mitochondrial DNA polymorphisms as risk factors for Parkinson’s disease and Parkinson’s disease dementia. Hum Genet. 2004;115:29–35. [DOI] [PubMed] [Google Scholar]
- 17.Wallace DC, Shoffner JM, Watts RL, et al. Mitochondrial oxidative phosphorylation defects in Parkinson’s disease. Ann Neurol. 1992;32:113–114. [DOI] [PubMed] [Google Scholar]
- 18.van der Walt JM, Nicodemus KK, Martin ER, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Walt JM, Dementieva YA, Martin ER, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–31. [DOI] [PubMed] [Google Scholar]
- 20.Shoffner JM, Brown MD, Torroni A, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. [DOI] [PubMed] [Google Scholar]
- 21.Giacchetti M, Monticelli I, De Biase L, et al. Mitochondrial DNA haplogroups influence the Friedreich’s phenotype. J Med Gen. 2004;41:293–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancuso M, Francesca LC, Rocchi A, et al. Could mitochondrial haplogroups play a role in sporadic amyotrophic lateral sclerosis? Neurosci Lett. 2004;371:158–162. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz-Pesini E, Lapena AC, Diez-Sanchez C, et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet. 2000;67:682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman S, Bezold R, Jaksch M, et al. Disease relevance of the so-called secondary Leber hereditary neuropathy mutations. Am J Hum Genet. 1997;60:1539–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann S, Bezold R, Jaksch M, et al. Wolfram (DIDMOAD) syndrome and Leber hereditary optic neuropathy (LHON) are associated with distinct mitochondrial DNA haplotypes. Genomics. 1997;39:8–18. [DOI] [PubMed] [Google Scholar]
- 26.Ross OA, McCormack R, Maxwell LD, et al. mt4216C variant in linkage with the mtDNA TJ cluster may confer a susceptibility to mitochondrial dysfunction resulting in an increased risk of Parkinson’s disease in the Irish. Exp Gerontol. 2003;38:397–405. [DOI] [PubMed] [Google Scholar]
- 27.Torroni A, Petrozzi M, D’Urbano L, et al. Haplotype and phylogenetic analyses suggest that 1 European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Gong JS, Zhang J, et al. Mitochondrial genotype is associated with longevity. Lancet. 1998;351:185–186. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova R, Leapage V, Charron D, et al. Mitochondrial genotype1 associated with French Caucasian centenarians. Gerontology. 1998;44:349. [DOI] [PubMed] [Google Scholar]
- 30.Ross OA, McCormack R, Curran MD, et al. Mitochondrial DNA polymorphism: its role in longevity of the Irish population. Exp Gerontol. 2001;36:1161–1178. [DOI] [PubMed] [Google Scholar]
- 31.Canter JA, Kallianpur AR, Parl FF, et al. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African American women. Cancer Res. 2005;65:8028–8033. [DOI] [PubMed] [Google Scholar]
- 32.Hulgan T, Haas DW, Haines JL, et al. Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS. 2005;19:1341–1349. [DOI] [PubMed] [Google Scholar]
- 33.Norris PR, Ozdas A, Cao H, et al. Cardiac uncoupling and heart rate variability stratify ICU patients by mortality: a study of 2088 trauma patients. Ann Surg. 2006;243:804–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris JA Jr, Eddy VA, Blinman TA, et al. The staged celiotomy for trauma. Issues in unpacking and reconstruction. Ann Surg. 1993;217:576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris JA Jr, Norris PR, Ozdas A, et al. Reduced heart rate variability: an indicator of cardiac uncoupling and diminished physiologic reserve in 1,425 trauma patients. J Trauma. 2006;60:1165–1173. [DOI] [PubMed] [Google Scholar]
- 36.Smeitink J, van den Heuvel L. Human mitochondrial complex I in health and disease. Am J Hum Genet. 1999;4:1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson BH. Human Complex I deficiency: clinical spectrum and involvement of oxygen free radicals in the pathogenicity of the defect. Biochim Biophys Acta. 1998;1364:271–286. [DOI] [PubMed] [Google Scholar]
- 38.Torroni A, Petrozzi M, D’Urbano L, et al. Haplotype and phylogenetic analyses suggest that 1 European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–11021. [PMC free article] [PubMed] [Google Scholar]
- 39.Howell N, Kubacka I, Halvorson S, et al. Phylogenetic analysis of the mitochondrial genomes from Leber hereditary optic neuropathy pedigrees. Genetics. 1995;140:285–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fauser S, Luberichs J, Besch D, et al. Sequence analysis of the complete mitochondrial genome in patients with Leber’s hereditary optic neuropathy lacking the three most common pathogenic DNA mutations. Biochem Biophys Res Commun. 2002;295:342–347. [DOI] [PubMed] [Google Scholar]
- 41.Canter JA, Kallianpur AR, Parl FF, et al. Response to letter from Mims et al. regarding mitochondrial DNA G10398A polymorphism and invasive breast cancer in African American women. Cancer Res. 2006;66:1880–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ioannidis JP, Trikalinos TA, Ntzani EE, et al. Genetic associations in large versus small studies: an empirical assessment. Lancet. 2003;361:567–571. [DOI] [PubMed] [Google Scholar]
- 43.Niemi AK, Moilanen JS, Tanaka M, et al. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet. 2005;13:166–170. [DOI] [PubMed] [Google Scholar]
- 44.Johnson SB, Lissaure M, Bochicchio GV, et al. Gene expression profiles differentiate between sterile SIRS and early sepsis. Ann Surg. 2007;245:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]