Abstract
The interaction of the chaperone SecB with ribosome-bound polypeptides that are in the process of elongation has been studied using an in vitro protein synthesis system. The binding is characterized by the same properties as those demonstrated for the binding of SecB to full-length proteins that are in nonnative conformation: it is readily reversible and has no specificity for the leader peptide. In addition, it is shown that the growing polypeptide chains must achieve a critical length to bind tightly enough to allow their isolation in complex with SecB. This explains the longstanding observation that, even when export is cotranslational, it begins late in synthesis. Furthermore, the required length is approximately the same as the length that defines the binding frame within denatured, full-length proteins bound to SecB.
SecB, a molecular chaperone in Escherichia coli, facilitates the export of a subset of periplasmic and outer membrane proteins by mediating a kinetic partitioning of polypeptides between the proper pathway of export and nonproductive pathways in the cytoplasm, which include folding and/or aggregation. Tetrameric SecB (68 kDa) binds precursor polypeptides and maintains them in a state that is lacking the stable conformation characteristic of the mature species, thereby allowing translocation across the cytoplasmic membrane (1). There are two reports by Watanabe and Blobel (2, 3) concluding that SecB specifically recognizes the leader of precursor polypeptides. However, numerous investigations carried out both in vivo and in vitro are in conflict with these studies and provide strong support for the conclusion that selective binding of precursors to SecB does not involve direct recognition of the leader sequence (4–15). The distinguishing feature of polypeptides that are ligands for SecB is that they are in a nonnative conformation. The mechanism of the recognition of a polypeptide as nonnative and the nature of the binding to SecB has been detailed using purified proteins. Characteristics of the interactions are as follows: (i) low specificity with high affinity (there is no apparent consensus in sequence and yet dissociation constants are in the submicromolar range; ref. 12); (ii) the binding is readily reversible, characterized by high rate constants both for association and dissociation (12, 13); and (iii) the binding frame mapped on two physiologic ligands is large, comprising a continuous stretch of at least 150 aa (14, 15).
A large proportion of export occurs after the polypeptides have been released from the ribosomes (16); thus, studies using purified proteins provide a good model for the posttranslational mode of binding to SecB. However, the export of some polypeptides begins while the chains are still undergoing elongation on the ribosome. For maltose-binding protein, 35% of export occurs cotranslationally (16). It seems that SecB is involved in both temporal modes of export, since fully elongated as well as nascent species of exported proteins can be isolated in complexes with SecB (11, 17) from exponentially growing cells. Here we report use of an in vitro protein synthesis system to characterize the interaction of SecB with ligands that are in the process of elongation on ribosomes. We show that interaction of SecB with nascent polypeptides in the complete absence of denaturant and its previously characterized interaction with full-length proteins after denaturation have the same properties. Thus the many studies of binding of SecB to proteins unfolded in vitro are relevant to the function of this chaperone in vivo. These results may be contrasted with a recent study in which two other chaperones, Hsc70 and TRiC, were shown to behave differently with full-length polypeptides and ribosome-bound nascent chains in vitro (18).
MATERIALS AND METHODS
Transcription in Vitro.
Plasmids pBAR43 (19) and pUZ226 (20), containing the malE gene with and without the leader sequence, respectively, were used to generate templates for transcription using PCR (21). The 5′ primer was designed so that in addition to the desired part of the coding sequence, the PCR products contained the T7 RNA polymerase promoter followed by a strong translation initiation sequence. The 3′ primers were designed so that translation in vitro would stop at a desired point in MalE. The reaction products in 0.5 ml were extracted twice with phenol:chloroform, precipitated with ethanol, washed twice with 75% ethanol, and dried at 60°C. The yield of DNA was typically 125 μg. Transcription of entire samples of DNA generated by PCR was carried out as described (21), but with the addition of 0.01% Triton X-100. The mRNA products were precipitated by addition of LiCl to 2.5 M, followed by incubation for 1 hr at −20°C, collected by centrifugation at 2000 × g for 10 min at 6°C, dissolved in 5 ml of sterile water, and reprecipitated with 2.5 M LiCl. The final pellet was dissolved in sterile water to a concentration of ≈10 μM and stored at −80°C. Each mRNA preparation was checked for uniformity of length by gel electrophoresis.
Translation in Vitro.
The concentrations of components of the in vitro translation system described (21) were optimized for maximal translation of each species of mRNA used. Reaction mixtures (0.1 ml) contained ≈100 pmol of 70S ribosomes and between 100 and 300 pmol of mRNA and were incubated as described at 37°C (10 min for the first incubation and 20 min for second incubation). Portions of the mixtures were then precipitated with trichloroacetic acid to estimate total [35S]methionine incorporated into polypeptides and for analysis of the radioactive products by SDS/PAGE. Other portions were applied to sucrose gradients or mixed with His6SecB (SecB with the following amino acyl extension on the amino terminus: Met-Arg-Gly-Ser-His-His-His-His-His-His-Gly-Ile-Arg) and applied to columns containing 0.2 ml of Ni2+-nitrilotriacetic acid (NTA) resin obtained from Qiagen (Chatsworth, CA).
Sucrose Gradient Centrifugation.
Translation mixtures were applied to sucrose gradients made by freezing and thawing 17.5% (wt/wt) sucrose (22) and centrifuged at 40,000 rpm for 1.5 hr at 6°C in the SW60 rotor (Beckman). Gradients were fractionated using an ISCO gradient fractionator that monitors absorbance at 254 nm. Trichloroacetic acid precipitates of portions of each fraction were used to estimate total radioactivity and for analysis of the radioactivity by SDS/PAGE. Other portions were applied to columns of Ni2+-NTA resin.
Chromatography on Ni2+-NTA Columns.
Samples of translation mixes or fractions from sucrose gradients, with or without added His6SecB, were applied to columns containing 0.2 ml of Ni2+-NTA resin equilibrated in 50 mM NaPO4 (pH 8.0), 0.3 M NaCl, and 5 mM MgCl2 and were followed by 1 ml of equilibration buffer. The columns were washed with 0.8 ml of 50 mM imidazole in 50 mM NaPO4 (pH 6.0), 0.3 M NaCl, and 5 mM MgCl2 and eluted with 3 ml of 0.5 M imidazole in the same buffer. All operations were carried out at 4°C. Portions of each fraction were precipitated with trichloroacetic acid for analysis by SDS/PAGE and determination of radioactivity.
Measurement of Radioactivity.
Radioactivity in individual bands on polyacrylamide gels or total radioactivity in single lanes was imaged and estimated with a PhosphorImager (Molecular Dynamics). The total [35S]methionine incorporated into polypeptides was estimated in trichloroacetic acid precipitates by scintillation counting.
Purification of Proteins.
SecB (9) and pure proteins added to the transcription and translation mixtures (21) were purified as described (9, 21). His6SecB was purified by using Ni2+ affinity chromatography from a strain that was a gift from C. A. Kumamoto (Tufts University School of Medicine).
RESULTS
The series of mRNAs used to program the translation system was obtained by the transcription of PCR-derived portions of the gene encoding maltose-binding protein and was designed so that translation would stop after elongation to aminoacyl residue 150, 280, or 370, which is the carboxyl-terminal residue of the full-length mature species of maltose-binding protein (21, 23). No stop codon was present in any of the constructs so that the polypeptides would remain on the ribosomes. In order that the ribosome would not run to the extreme 3′ end of the RNA, in every case two codons for cysteine followed by 20 adenine residues were added to the mRNA immediately following the last codon for the amino acid to be translated from the maltose-binding protein. Maltose-binding protein contains no cysteine; thus omission of cysteine from the mixture of amino acids provided in the in vitro system allowed elongation up to, but not through, the cysteine codons. Two series of mRNA were made: one series included the codons for the leader sequence, which consists of 26 aa at the amino terminus (conventionally numbered −26 to −1) and the other series had the codon for the first aminoacyl residue of the mature species immediately following the codon for the initiating methionine.
Characterization of Ribosome-Bound Nascent Polypeptides.
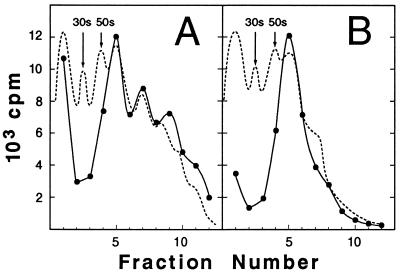
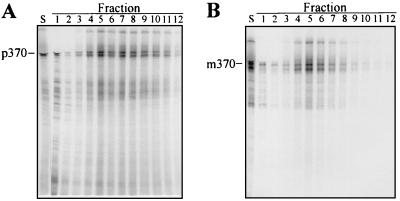
As expected, the polypeptides synthesized in the system programmed with these mRNAs and radiolabeled with [35S]methionine remained associated with the ribosomes. Whether the mRNA used encoded precursor species (Fig. 1A, precursor encoded to amino acid 370, designated p370) or mature species (Fig. 1B, mature protein encoded to amino acid 370, designated m370), the fractionation of reaction mixtures by sucrose gradient centrifugation gave similar results. The great majority (>80%) of the labeled polypeptides sedimented into the gradient and were distributed in a pattern that corresponded to the distribution of the ribosomes, as determined by absorbance at 254 nm. Inspection of the A254 profiles (Fig. 1) as well as labeling with [3H-formyl]methionine (data not shown) showed that, routinely, between 10% and 30% of the ribosomes were active in initiating protein synthesis in the presence of excess mRNA. The mRNA for the precursor species consistently gave a higher level of initiation. Analysis of the radiolabeled polypeptide products by SDS/PAGE followed by imaging with a PhosphorImager showed that, in addition to the full-length species of precursor and mature proteins, a second prominent distinct molecular weight species of each was present just below the full-length species on the gel (Fig. 2 A and B). These shorter polypeptide species might represent the nascent chain carried on the second ribosome of a polysome; however, since the ratio of the full-length to the shorter species was constant whether the fraction examined contained monosomes, disomes, or polysomes composed of higher numbers of ribosomes, this seems unlikely. It is more probable that the distinct molecular weight species represent pauses in synthesis, especially since pauses in synthesis of maltose-binding protein have been demonstrated both in vivo and in vitro (24). Some of the distinct species of lower molecular weight seen among the products of the precursor mRNA are likely to be the result of premature termination, since they did not sediment into the gradient with the ribosomes.
Figure 1.
Products of in vitro translation remain associated with ribosomes. After incubation, translation mixtures were applied to sucrose gradients and centrifuged. The absorbance at 254 nm (dotted line) and the 35S in trichloroacetic acid precipitates of portions of the individual fractions (•) are shown. Translation mixtures contained (A) mRNA for p370 and (B) mRNA for m370.
Figure 2.
Analysis of ribosome-associated translation products. A portion of each fraction of the gradients shown in Fig. 1 was trichloroacetic acid-precipitated and subjected to SDS/PAGE on a 14% acrylamide gel. The 35S-labeled products were displayed using a PhosphorImager. Lane S contains a portion of the sample applied to the gradient.
Binding of Nascent Chains to SecB.
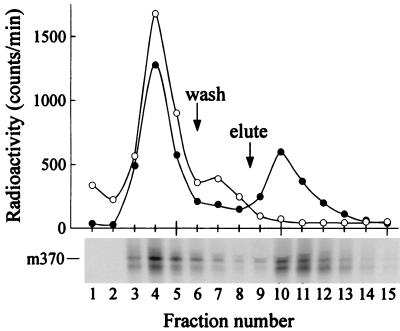
Binding of nascent polypeptides to SecB could be demonstrated using SecB, engineered to carry a His6 tag at the amino terminus, so that it would bind with high affinity to a resin that contains chelated Ni2+. Following sucrose gradient centrifugation of the protein synthesis reaction mixture programmed with the mRNA for the mature full-length species, a sample (Figs. 1B and 2B, fractions 4 and 6 pooled) that contained ribosome-bound radiolabeled polypeptides was mixed with His6SecB and applied to a column containing the Ni2+ resin (Fig. 3, •). Polysomes bound to the His6SecB via their nascent polypeptides should be retained by the Ni2+ resin, while those not in complex with SecB should flow through. Of the radiolabeled polypeptides, 35% were retained and were subsequently eluted by 0.5 M imidazole, which competes with histidine for binding the Ni2+ and releases the His6SecB. When the ribosome-bound polypeptides were applied to the column without addition of His6SecB (Fig. 3, ○), all of the radioactivity was recovered either in the flow-through or in the 0.05 M imidazole wash, which releases material that is bound weakly to the resin. For the experiment shown, His6SecB was added to the sample to give a concentration of 1.5 μM SecB tetramer. When the concentration of His6SecB was varied from 0.07 μM to 1.5 μM, there was no increase in the amount of radiolabeled polypeptides bound above 0.37 μM SecB. It was demonstrated that addition of a wash of 1 M potassium acetate following the 0.05 M imidazole wash did not release radioactivity from columns loaded with polysomes bound to His6SecB and that the polypeptides eluted together with His6SecB by 0.5 M imidazole were still bound to polysomes, since they sedimented with ribosomes upon sucrose gradient centrifugation (data not shown).
Figure 3.
Ribosome-associated polypeptides bind SecB. Portions of pooled fractions 4 and 6 from the gradient shown in Fig. 1B, with (•) and without (○) His6SecB added, were applied to columns containing 0.2 ml of Ni2+-NTA resin and were followed by 1 ml of equilibration buffer. The columns were washed with 0.8 ml of 50 mM imidazole in wash buffer (wash) and eluted with 3 ml of 0.5 M imidazole in wash buffer (elute). Portions of each fraction from the chromatogram of the sample containing His6SecB were precipitated for determination of radioactivity and analysis by SDS/PAGE. The position of the full-length mature species is indicated (m370).
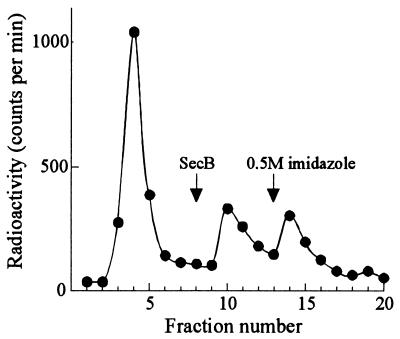
It has been shown previously that the binding of full-length nonnative proteins to SecB, although of high affinity, is characterized by high rate constants for both association and dissociation—i.e., it is readily reversible (12, 13). This is also the case for the interaction of SecB with the ribosome-bound nascent chains. When SecB, which did not have a His6 tag, was applied to a column that had been loaded with polysomes bound to His6SecB, more than half of the bound radiolabeled polypeptides were eluted with the SecB (Fig. 4). Thus, there is a rapid conversion between the bound and free states of the nascent chains, allowing them to bind to, and elute with, the competing SecB.
Figure 4.
Binding of ribosome-associated polypeptides to SecB is reversible. Column chromatography of a gradient fraction containing ribosome-bound polypeptides (m370) to which His6SecB (0.7 μM) was added was carried out as described for Fig. 3, except that following the 0.8-ml wash, the column received in the following order: 0.1 ml of SecB (1 mg/ml in wash buffer), 1 ml of wash buffer, then 3 ml of elution buffer.
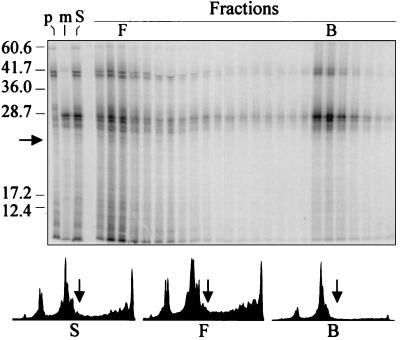
The presence of a leader sequence is not a requirement for binding of full-length polypeptides by SecB either in vitro (7–10) or in vivo (4–6, 11). As shown in Fig. 5, this is also the case for polypeptides bound to ribosomes. The products of mRNA encoding the full-length precursor (p370; Fig. 5, lane p) were mixed with the products of the mRNA encoding the mature species ending at residue 280 (m280; Fig. 5, lane m) so that the precursor and mature species could be easily resolved by molecular weight. The mixture (Fig. 5, lane S) was incubated with His6SecB and applied to the Ni2+ resin column. The radiolabeled polypeptides contained in the sample applied to the column as well as those in each of the fractions of the column chromatography were separated according to size by SDS/PAGE and detected using a PhosphorImager (Fig. 5 Upper). Quantification of the images obtained showed that the same proportion (≈25%) of each species was bound and eluted by 0.5 M imidazole (data in Fig. 5), indicating that even when undergoing elongation, polypeptides with and without the leader bind equally well.
Figure 5.
Leader peptide is not required for binding of nascent polypeptides to SecB. Separate mixtures, programmed with mRNA for p370 (lane p) and m280 (lane m), were combined after translation (lane S) and His6SecB (0.5 μM) was added. Column chromatography was carried out as described for Fig. 3, except that the volume of the wash was 4 ml. Fractions were analyzed by SDS/PAGE. The intensity profiles of the radioactivity in the lanes S (sample, total applied), F (free, not retained by the His6SecB on the column), and B (bound, retained on the column and eluted by 0.5 M imidazole), were generated using imagequant software version 4.2 (Molecular Dynamics). The arrow indicates the position for polypeptides of a molecular weight of 23,000, as calculated using the molecular weight markers shown.
Length Requirement for Binding to SecB.
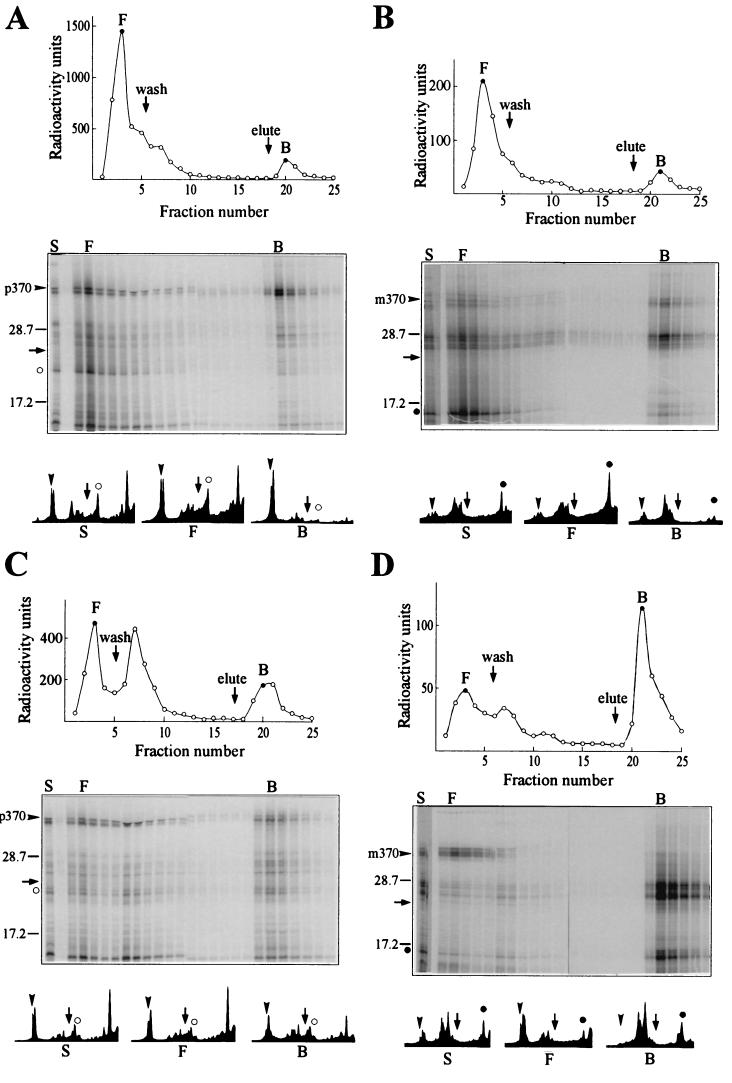
The population of polypeptides that is recovered in the bound fraction shows an enrichment for those of molecular weight greater than ≈23,000, as is clearly seen by a comparison of the size distribution of polypeptides in the sample applied with that of polypeptides in the material that flows through and with that of polypeptides recovered in the bound fraction [Fig. 5 Lower; compare intensity profiles of sample (S), free (F), and bound (B)]. With the exception of one product of molecular weight <10,000 derived from the mRNA for the p370 species, most of the radioactivity in the low molecular weight region of the gel is present as a diffuse smear, as is expected, since the population of elongating polypeptides is heterodisperse with respect to length. Thus to examine binding of nascent polypeptides as a function of length, we programmed separate synthesis mixtures with a single mRNA species encoding either a full-length species (p370 or m370), a species ending at amino acid 280 (m280), or a species terminating at residue 150 (p150 or m150). Since the leader sequence contains three methionine residues (23), precursor polypeptide chains undergoing elongation that were shorter than 150 aa were easily detectable when radiolabeled with [35S]methionine. However, the first methionine outside of the leader is at position 145. Thus a polypeptide of 145 residues in length is the shortest polypeptide translated from the mature mRNA that will be labeled. To facilitate detection, the mRNA for m150 was translated using [35S]methionine at a specific activity three times as high as that used for the other species. The translated products of mRNAs encoding precursor species were pooled separately from those products derived from mRNAs for the mature species and each mixture was assessed for binding to SecB. It was clear that for both the species with (Fig. 6A) and the species without the leader (Fig. 6B), the population of polypeptide chains that bound to His6SecB tightly enough to allow retention on the column was enriched for chains elongated further than amino acid residue 150 in the mature sequence (m150, solid circles; 176 aa for p150, open circles). Quantifications of the intensity profiles in Fig. 6 show that, whereas in the population of polypeptides not retained on the column (Fig. 6 A and B, profiles F), approximately half of the species were below a molecular weight of 23,000 [Fig. 6 A (59%) and B (47%)], in the population of polypeptides retained (Fig. 6 A and B, profiles B), <20% of the species were below a molecular weight of 23,000 [Fig. 6 A (19%) and B (17%)]. Since the concentration of His6SecB when it is tethered to the column is not known, we cannot quantitatively determine the affinities of the different species. Nevertheless, the large difference in recovery of polypeptides in the bound fraction that occurs abruptly between lengths of 150 and 200 aa (Figs. 5 and 6 A and B) must reflect a large difference in affinity. It should be noted that this preferential binding of chains of a molecular weight of >23,000 accounts for the difference in percentage of radiolabeled polypeptides recovered in the bound fraction between the experiments displayed in Figs. 3 and 4 (35% and 45% bound, respectively), in which only the full-length mRNAs were used, and those in Fig. 6 A and B (7% and 13% bound, respectively), in which a large proportion of the radioactivity was in low molecular weight species.
Figure 6.
Size requirement of SecB ligands. Separate reaction mixtures were programmed with mRNAs for p370, p150, m370, m280, and m150. After incubation, the mixtures containing p370 and p150 were pooled (A and C), as were those containing m370, m280, and m150 (B and D). Half of each pool was incubated with (C and D) or without (A and B) 0.4 mM puromycin for 5 min at 37°C, mixed with His6SecB (0.8 μM), and subjected to column chromatography and analysis, as described for Fig. 5. The positions of p150 (○) and m150 (•) are indicated on the intensity profiles of lanes S, F, and B. The arrow in the lower half of each figure marks molecular weight of 23,000, as in Fig. 5.
Nascent chains elongated to 150 residues would have between 24 residues (if extended) and 56 residues (if helical) within the ribosomal channel, which is 85 Å long (25), and thus only 94–126 residues would be accessible to SecB. This length might be too short to fill the binding site on SecB, since it has been determined that the binding frame within two natural ligands, galactose-binding protein and maltose-binding protein, extends over a contiguous sequence of between 150 and 170 aa (14, 15). An alternative explanation is that the short nascent chains are in complex with other proteins and not accessible to SecB. That the former explanation is the more likely is indicated by recovery in the bound fraction of the polypeptide chains elongated to amino acid 150 after incubation of polysomes with puromycin to release the nascent chains from the ribosomes [Fig. 6 C (p150; open circles) and D (m150; solid circles)]. Not only are shorter chains recovered among the bound polypeptides, but after release from the ribosome the longest mature species—i.e., the full-length m370 and the distinct species attributed to a pause in synthesis—no longer bind (Fig. 6D). It is well documented that SecB does not bind to stably folded, native maltose-binding protein (6), and it is likely that upon release from the ribosome, the full-length mature protein and the species that lacks only a few carboxyl-terminal amino acids fold and thereby escape binding.
DISCUSSION
The results presented here contrast with those in a recent study of two other chaperones, Hsc70 and TRiC (18), by establishing that the principles governing the interaction of SecB with nascent polypeptides on the ribosome are the same as those underlying the binding of full-length, purified polypeptides, rendered nonnative by incubation with denaturant. SecB does not select its ligands by specifically binding to the leader peptide. Nascent polypeptides were shown to bind SecB whether or not they contained a leader peptide. The binding to polypeptides that are tethered to the ribosome is readily reversible as was previously shown to be the case with free, full-length, physiologic ligands (11, 12) as well as with the model ligand, nonnative bovine pancreatic trypsin inhibitor (12, 13). Thus there is no need to invoke interaction with other cellular components such as ATP or SecA to cause release of the ligand from SecB. Transfer of the ligand from SecB to SecA, a component of the export translocase, will occur as long as the affinity for the ligand is higher.
The observation that polypeptides must acquire a length of ≈200 aa if nascent or 150 aa if released from the ribosome to bind tightly enough to be isolated in complex with SecB is consistent with the finding that the binding of SecB to the full-length, denatured ligands, maltose-binding protein and galactose-binding protein, protects 170 aa and 150 aa, respectively, of these ligands (14, 15). It has been proposed that the binding site on SecB comprises multiple subsites that each bind flexible stretches of polypeptide ligand and that the high affinity observed for binding of full-length ligands results from the simultaneous occupation of these subsites. Thus a short peptide that binds at only one subsite would be expected to have a lower affinity than a long ligand, which could bind at more than one of the subsites. In addition, it is proposed that upon simultaneous occupation of several subsites, SecB undergoes a conformational change, exposing a hydrophobic site that also interacts with the ligand (26). Within the framework of this model the abrupt increase in affinity for nascent chains that have >150 aa accessible to SecB can be explained in two ways. The 150 aa may fill the entire binding site on SecB, even though the primary sequence of residues 1–150 is not the same as that of the defined binding frame within maltose-binding protein, which extends from residue 77 to residue 240. The precise sequence may not be crucial, since the sequence of the 150 aa that defines the binding frame within galactose-binding protein has no obvious similarity to the sequence of the binding frame within maltose-binding protein. However, it should be noted that amino acid residue 150 lies in the middle of the defined binding frame. Therefore there is an alternative possibility that the nascent chains are bound via the same sequence as are full-length proteins, but only half of the available site on SecB is occupied. The affinity would be lower than if the entire site were filled and yet it might be sufficiently high to allow retention on the column. Whichever explanation is correct, the requirement for a molecular weight of 23,000 to render the nascent chain a good ligand for SecB explains the observation that even when cotranslational, export of maltose-binding protein begins only after the polypeptide has reached a molecular weight of 33,000 (16).
The data acquired from these studies of the interaction of SecB with nascent polypeptides together with that from previous work carried out both in vitro with full-length protein (7–10, 12–15) and in vivo (4–6, 11, 16, 17) can all be rationalized in one consistent model. SecB facilitates export by binding polypeptides rapidly, before they acquire stable structure. Once folded, a precursor polypeptide is no longer capable of being exported despite the presence of the leader sequence. Although binding is of high affinity, it is readily reversible, because both the kon and koff are high. Interaction with SecB is readily reversible both for ribosome-bound polypeptides as well as for those polypeptides that interact posttranslationally, implying that the parameters governing binding are similar. For those chains that are translocated before elongation is completed, the primary role of SecB might be considered to be one of promoting entry into the export pathway by its affinity for SecA. However, as shown here, a species that is missing ≈30 residues from the carboxyl terminus acquires a conformation that SecB cannot bind, and since cotranslational transfer begins only after elongation of ≈300 of the 370 residues in the mature species (16), a significant proportion of chains undergoing elongation of the final 70 residues might be capable of folding. Therefore, there should be a kinetic partitioning between folding and export for those polypeptides elongated sufficiently to be capable of acquiring a conformation that precludes SecB binding. Clearly, there is no question that the population of polypeptides that is translocated after elongation is complete (for maltose-binding protein, approximately 65%) is subject to a kinetic partitioning between folding and export.
Acknowledgments
We thank Prof. C. G. Kurland for providing us with laboratory space and facilities in the Department of Molecular Biology at the University of Uppsala. We are grateful to R. Karimi, V. Dincbas, and N. Bilgin of the Department of Molecular Biology at the University of Uppsala for advice and preparation of factors. We thank V. F. Smith for purifying His6SecB using Ni2+ affinity chromatography. This work was supported by National Institutes of Health Grant GM 29798 to L.L.R., by a grant from the Swedish Research Council for Engineering Sciences to M.E., and by a grant from the Swedish Cancer Society.
Footnotes
Abbreviations: His6SecB, SecB with the following amino acyl extension on the amino terminus: Met-Arg-Gly-Ser-His-His-His-His-His-His-Gly-Ile-Arg; NTA, nitrilotriacetic acid.
References
- 1.Randall L L, Hardy S J S. Trends Biochem Sci. 1995;20:65–69. doi: 10.1016/s0968-0004(00)88959-8. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe M, Blobel G. Proc Natl Acad Sci USA. 1989;86:2728–2732. doi: 10.1073/pnas.86.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe M, Blobel G. Proc Natl Acad Sci USA. 1995;92:10133–10136. doi: 10.1073/pnas.92.22.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier D N, Bankaitis V A, Weiss J B, Bassford P J., Jr Cell. 1988;53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 5.Liu G, Topping T B, Randall L L. Proc Natl Acad Sci USA. 1989;86:9213–9217. doi: 10.1073/pnas.86.23.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gannon P M, Li P, Kumamoto C A. J Bacteriol. 1989;171:813–818. doi: 10.1128/jb.171.2.813-818.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecker S, Lill R, Ziegelhoffer T, Georgopoulos C, Bassford P J, Jr, Kumamoto C A, Wickner W. EMBO J. 1989;8:2703–2709. doi: 10.1002/j.1460-2075.1989.tb08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss J B, Bassford P J., Jr J Bacteriol. 1990;172:3023–3029. doi: 10.1128/jb.172.6.3023-3029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randall L L, Topping T B, Hardy S J S. Science. 1990;248:860–863. doi: 10.1126/science.2188362. [DOI] [PubMed] [Google Scholar]
- 10.de Cock H, Overeem W, Tommassen J. J Mol Biol. 1992;224:369–379. doi: 10.1016/0022-2836(92)91001-6. [DOI] [PubMed] [Google Scholar]
- 11.Khisty V J, Randall L L. J Bacteriol. 1995;177:3277–3282. doi: 10.1128/jb.177.11.3277-3282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy S J S, Randall L L. Science. 1991;251:439–443. doi: 10.1126/science.1989077. [DOI] [PubMed] [Google Scholar]
- 13.Fekkes P, den Blaauwen T, Driessen A J M. Biochemistry. 1995;34:10078–10085. doi: 10.1021/bi00031a032. [DOI] [PubMed] [Google Scholar]
- 14.Topping T B, Randall L L. Protein Sci. 1994;3:730–736. doi: 10.1002/pro.5560030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khisty V J, Munske G R, Randall L L. J Biol Chem. 1995;270:25920–25927. doi: 10.1074/jbc.270.43.25920. [DOI] [PubMed] [Google Scholar]
- 16.Josefsson L-G, Randall L L. Cell. 1981;25:151–157. doi: 10.1016/0092-8674(81)90239-7. [DOI] [PubMed] [Google Scholar]
- 17.Kumamoto C A, Francetic O. J Bacteriol. 1993;175:2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frydman J, Hartl F U. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 19.Rasmussen A, MacGregor C H, Ray P H, Bassford P J., Jr J Bacteriol. 1985;164:665–673. doi: 10.1128/jb.164.2.665-673.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss J B, MacGregor C H, Collier D N, Fikes J D, Ray P H, Bassford P J., Jr J Biol Chem. 1989;264:3021–3027. [PubMed] [Google Scholar]
- 21.Pavlov M Y, Ehrenberg M. Arch Biochem Biophys. 1996;328:9–16. doi: 10.1006/abbi.1996.0136. [DOI] [PubMed] [Google Scholar]
- 22.Randall L L, Hardy S J S. Methods Enzymol. 1983;97:70–76. doi: 10.1016/0076-6879(83)97120-3. [DOI] [PubMed] [Google Scholar]
- 23.Duplay P, Bedouelle H, Fowler A, Zabin I, Saurin W, Hofnung M. J Biol Chem. 1984;259:10606–10613. [PubMed] [Google Scholar]
- 24.Randall L L, Josefsson L-G, Hardy S J S. Eur J Biochem. 1980;107:375–379. doi: 10.1111/j.1432-1033.1980.tb06039.x. [DOI] [PubMed] [Google Scholar]
- 25.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata R K, Agrawal R K. Nature (London) 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 26.Randall L L. Science. 1992;257:241–245. doi: 10.1126/science.1631545. [DOI] [PubMed] [Google Scholar]