Abstract
The cationic amino acid transporter, Cat-1, is a high affinity transporter of the essential amino acids, arginine and lysine. Expression of the cat-1 gene increases during nutritional stress as part of the adaptive response to starvation. Amino acid limitation induces coordinate increases in stability and translation of the cat-1 mRNA, at a time when global protein synthesis decreases. It is shown here that increased cat-1 mRNA stability requires an 11 nucleotide AU-rich element within the distal 217 bases of the 3′-untranslated region. When this 217-nucleotide nutrient sensor AU-rich element (NS-ARE) is present in a chimeric mRNA it confers mRNA stabilization during amino acid starvation. HuR is a member of the ELAV family of RNA-binding proteins that has been implicated in regulating the stability of ARE-containing mRNAs. We show here that the cytoplasmic concentration of HuR increases during amino acid starvation, at a time when total cellular HuR levels decrease. In addition, RNA gel shift experiments in vitro demonstrated that HuR binds to the NS-ARE and binding was dependent on the 11 residue AU-rich element. Moreover, HuR binding to the NS-ARE in extracts from amino acid-starved cells increased in parallel with the accumulation of cytoplasmic HuR. It is proposed that an adaptive response of cells to nutritional stress results in increased mRNA stability mediated by HuR binding to the NS-ARE.
Mammalian cells have an adaptive response to limited nutrient supply (reviewed in Refs. 1, 2, and 3). This response promotes expression of genes essential for cell survival at a time when global protein synthesis decreases (2, 3). One of the genes whose expression is induced during nutrient limitation is cat-1,1 the high affinity transporter for the essential amino acids Arg and Lys (4, 5). We previously showed that the increased Cat-1 protein expression during amino acid deprivation is due to increased mRNA levels and enhanced translation of the mRNA (6–11). We also showed that the increased mRNA levels are mainly due to increased mRNA stability mediated by sequences within the 4.5-kb 3′-UTR of the 7.9-kb cat-1 mRNA (10, 12). It is shown here that an 11-residue AU-rich element in the distal 217 bases of the 3′-UTR is required for increased mRNA stability during amino acid starvation. Moreover, this element functions in both the cat-1 mRNA and in a chimeric mRNA.
To begin to address the mechanism of mRNA stabilization during amino acid starvation, the interaction of the cat-1 mRNA with HuR protein was examined. HuR is a ubiquitously-expressed member of the embryonic lethal abnormal vision (ELAV) family of RNA-binding proteins (reviewed in Refs. 13, 14). This protein is found in both the nucleus and cytoplasm. It shuttles between these two compartments, and it may play a role in mRNA export from the nucleus (15, 16). HuR has been shown to bind to AU-rich elements in many mRNAs and to play a role in the stabilization of these mRNAs (13). Finally, HuR functions are regulated. Several cellular stresses, including UV irradiation, heat shock, and actinomycin D shift HuR from the nucleus to the cytoplasm (17–19). Furthermore, changes in mRNA stability caused by these stresses may be mediated by HuR.
We show that HuR binds to the AU-rich element in the 3′-UTR of the cat-1 mRNA. Moreover, starvation causes HuR accumulation in the cytoplasm. Finally, the ability of cytoplasmic HuR to bind to the cat-1 AU-rich element shows a transient increase during amino acid starvation and these changes match the time course of cat-1 mRNA accumulation in vivo. Taken together, these results suggest that HuR may play a role in stabilizing the cat-1 mRNA during amino acid deprivation.
EXPERIMENTAL PROCEDURES
Tissue Culture Cells and Transfections
C6 rat glioma cells were maintained in 10% fetal bovine serum-supplemented Dulbecco’s modified Eagle’s medium (DMEM)/F-12 medium. Amino acid-fed cells were cultured in dialyzed fetal bovine serum-containing DMEM/F12 (Fed), whereas cells were amino acid depleted by culturing in dialyzed fetal bovine serum-supplemented Krebs-Ringer bicarbonate (KRB) buffer (Starved). Cells were transfected with the tetcat-1 or pCAT plasmids along with an expression vector containing the neo gene, and stable lines were selected in 0.1% G418.
Expression Vectors
The ptetcat-1 vector contains the cat-1 cDNA (+1 to 6453, Fig. 1A) under the transcriptional control of the tetracycline-inducible promoter (10). The first 80 bases of the tetcat-1 mRNA are from the tet promoter. The ptetcat-1/Δ1, Δ2, and Δ3 expression vectors (Fig. 1) were generated by deleting sequences from the 3′-UTR of wild type ptetcat-1 (10). ptetcat-1/Δ1 was generated by deleting the StuI/NdeI fragment (2277–3470). ptetcat-1/Δ2 was generated by deleting the NdeI/EcoRV fragment (3470–4786). ptetcat-1/Δ3 was generated by deleting the HindIII fragment (4310–5894). ptetcat-1/Δ4 and ptetcat-1/Δ5 were generated in two steps using PCR-based methods. First, a BstEII restriction site was introduced at position 6445 between the polyadenylation signal, AATAAA (position 6432) and the polyadenylation site (position 6453). mRNA from this vector accumulated during amino acid deprivation similarly to the tetcat-1 mRNA (Fig. 1D, con). To generate ptetcat-1/Δ4 -ptetcat-1/Δ6, the 759-bp BstEII fragment (5695–6453) was replaced with truncated BstEII fragments containing residues 5695–6078 (ptetcat-1/Δ4) and 5695–6236 (ptetcat-1/Δ5). ptetcat-1/Δ4-AUmut was generated by mutating residues 6268–6279 of the cat-1 cDNA sequence, (TATTTTATTTTA) to GATGGATGGTA in ptetcat-1/Δ4. ptetcat-1/Δ6 was generated by replacing the BstEII fragment (5695–6453), which contains the 3′-end of the cat-1 gene, with the SV40 polyadenylation signal sequence. pCAT was the pCAT®3 control vector from Promega. pCAT/UTR was constructed by inserting the BstEII fragment from ptetcat-1 into the XbaI/HpaI sites of pCAT by blunt-end ligation. This fragment contains residues 5965–6453 from the cat-1 3′-UTR and the SV40 late polyadenylation region.
Fig. 1. An AU-rich element within the 3′-UTR mediates increased cat-1 mRNA levels during amino acid starvation.
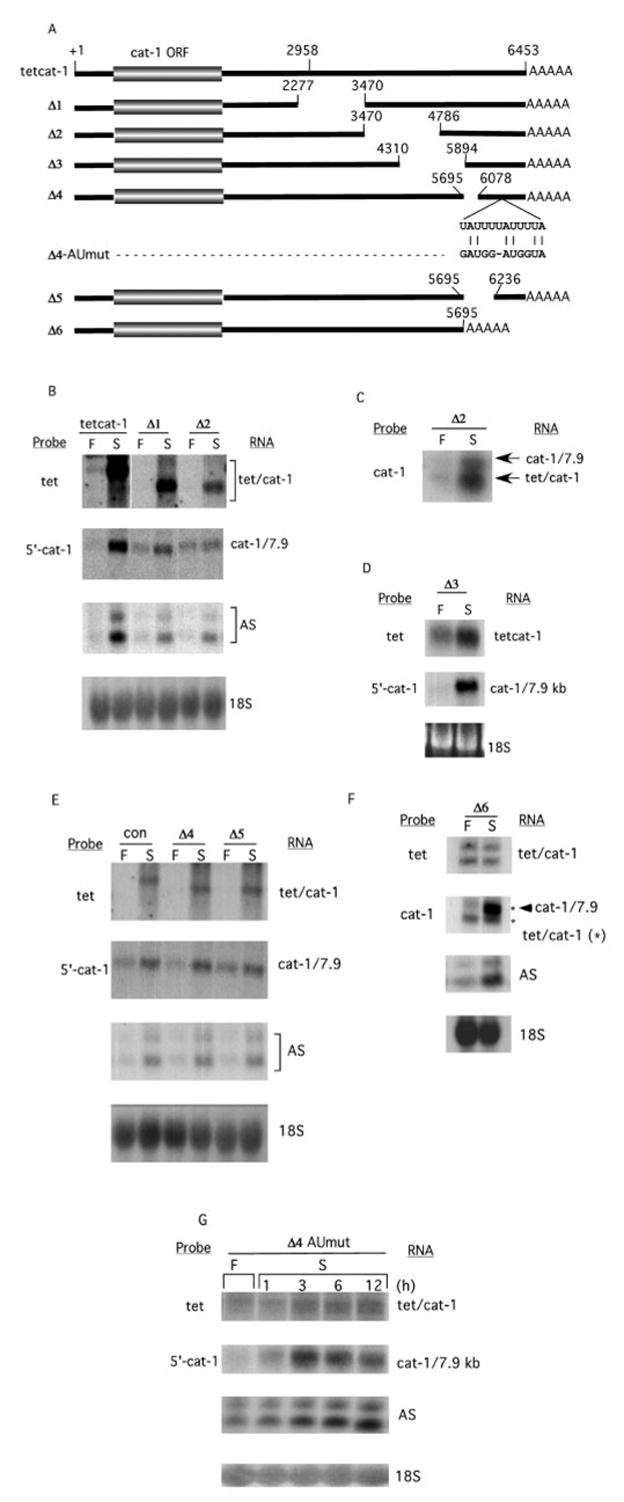
A, cDNAs used to study mRNA stability. +1, indicates the beginning of the cDNAs, which contain 79 bases of cat-1 5′-UTR. Gaps indicate deleted sequences. For the Δ4-AUmut construct, the wild type sequence (residues 6268–6279) and the sequence of the mutated construct are shown. B–G, Northern blot analysis of RNA (15 μg except for F, 40 μg) isolated from C6 cells stably transfected with the DNAs in A. The control (Con) plasmid in E is ptetcat-1 that contains the BstEII site. RNA was isolated from cells incubated for 4 h (or as indicated) in either amino acid-containing (F) or amino acid-depleted (S) media. Probes used include tet, which specifically detects tetcat-1 mRNAs; 5′-cat-1, which specifically detects endogenous cat-1 mRNA; and cat-1, which detects both endogenous cat-1 and tetcat-1 mRNAs. Specific probes were also used to detect asparagine synthase mRNAs (AS) and 18 S ribosomal RNA (18S), all as described under “Experimental Procedures.” In panel F, the migration positions of tetcat-1 RNAs are indicated by an asterisk.
Northern and Western Blot Analyses
RNAs were detected by Northern blotting using the following 32P-labeled DNA probes: tet is a 157-bp KpnI fragment from ptetcat-1 that contains the first 70 bp of the tetcat-1 RNA (10). This probe hybridizes to the tetcat-1 RNAs but not the endogenous cat-1 mRNA. 5′-cat-1 is a 0.1-kb fragment from the first exon of the cat-1 gene (6) that hybridizes to endogenous cat-1 mRNA but not the tetcat-1 RNAs. cat-1 was the entire cat-1 cDNA. This probe detects both endogenous cat-1 and tetcat-1 RNAs.
Asparagine synthase (AS) was detected using a 900-bp fragment of the AS cDNA (20). 18 S ribosomal RNA was detected with a 5.8-kb fragment containing the 18 S mouse ribosomal DNA (21). RNAs from pCAT and pCAT/UTR were detected with a probe from the chloramphenicol acetyltransferase DNA.
Western blot analysis of HuR, AUF1, and actin proteins were carried out using the appropriate antibodies. The HuR antibody, a generous gift of H. M. Fourneaux, has been described previously (22). The AUF1 antibody was generated as follows: a cDNA expressing the 37-kDa isoform of AUF1 (provided by Dr. Gary Brewer, UMDNJ), was sub-cloned into a T7-dependent bacterial expression system. p37AUF1 protein was produced in Escherichia coli and used to raise a rabbit polyclonal antibody. The antibody was affinity purified. The anti-actin antibody (H-196) was from Santa Cruz Biotechnology.
Cell Fractionation
Nuclear and cytoplasmic extracts were generated as follows: Cells (108) were harvested in phosphate-buffered saline and pelleted and suspended in lysis buffer (10 mM HEPES pH 7.9, 20 mM KCl, 3 mM MgCl2, 0.5% Nonidet P-40, 5% glycerol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride). Cells were kept on ice for 10 min, during which time they were sheared 5 times through a 25-gauge needle. Lysates were centrifuged at 6,000 rpm for 10 min, and the supernatant was saved as cytoplasmic lysate. The pellet was suspended in nuclear lysis buffer (20 mM HEPES pH 7.9, 0.225 M NaCl, 1 mM EDTA, 3 mM MgCl2, 0.5% Nonidet P-40, 10% glycerol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride), sheared by passing through a 27-gauge needle and kept on ice for 15 min. Nuclear extracts were obtained by collecting the supernatant after a 14,000 rpm spin for 10 min at 4°. Extracts were kept at −80 °C until use.
RNA Electrophoretic Mobility Shift Assays (EMSA) and UV Cross-linking
32P-labeled RNA probes (specific activity 1.5 × 107 cpm/μg) were generated by in vitro transcription of PCR-generated DNA templates containing the T7 promoter (Ambion), followed by gel purification. The wild-type RNA had the sequence of residues 6232–6308 of the cat-1 cDNA. The AUmut RNA had GAUGGAUGGUA substituted for UAUUUUAUUUUA beginning at residue 6268 of the rat cat-1 cDNA. RNA binding reactions were performed by incubating cell lysates (0.6 mg of protein/ml) in the presence of 5 mg/ml heparin for 15 min at room temperature, followed by the addition of [32P]RNA (105 cpm/15-μl reaction) for an additional 20 min. Antibodies were then added for 1 h on ice. Complexes were resolved on 5% non-denaturing polyacrylamide gels. To analyze proteins bound to the RNA, samples were irradiated for 30 min on ice in a Stratalinker UV Crosslinker (Stratagene), followed by incubation with RNase T1 (0.2 units) and RNase A (20 μg) at 37 °C for 30 min. Samples were analyzed by sodium dodecyl sulfate electrophoresis on 7.5% gels.
Immunocytochemistry
Cells were plated on Lab-Tec chamber slides 36 h before the experiment. Cells were fed with fresh medium for 12 h and then incubated in amino acid-containing or amino acid-depleted medium for the indicated times. After treatment, cells were fixed in freshly-prepared 4% paraformaldehyde for 20 min at room temperature. Slides were then quenched in 25 mM glycine in phosphate-buffered saline and blocked for 1 h in 2% bovine serum albumin, 5% fish gelatin, and 0.02% saponin. Samples were incubated with antibody to HuR (20 μg/ml) in blocking solution overnight at 4 °C. Slides were washed three times for 15 min in blocking buffer and then incubated with Oregon Green-conjugated anti-mouse IgG (1:300) (Molecular Probes, Eugene, OR) for 1 h in blocking buffer. Slides were finally washed three times for 15 min in blocking buffer and twice for 5 min in phosphate-buffered saline. Slides were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Cells were examined with either a BioRad confocal microscope or a Nikon microscope. At least four separate preparations were examined for each condition. Nonspecific staining, assessed by incubating the cells in the absence of primary antibody, was negligible.
RESULTS
An AU-rich Element in the 3′-UTR of the cat-1 mRNA Confers Increased mRNA Stability during Amino Acid Starvation
mRNAs of ~7.9 and ~3.4 kb are synthesized from the cat-1 gene. These mRNAs result from polyadenylation at two sites within the 3′-UTR (10). Nuclear run-off studies have shown that the transcription rate of the cat-1 gene does not change during amino acid limitation (10). Furthermore, we have previously shown that the stability of the 7.9 kb but not the 3.4 kb cat-1 mRNA increases in amino acid-depleted cells (10). These findings suggested that sequences which regulate mRNA stability during amino acid starvation are contained within the 3.5 kb at the 3′-end of the 7.9 kb cat-1 mRNA that is not present in the 3.4 kb mRNA (10).
In order to identify the regulatory cis-mRNA sequences, a series of cDNAs with truncations in the 3′-UTR of the 7.9 kb mRNA were generated (Fig. 1A). These cDNAs were linked to a tetracycline-inducible promoter in an expression vector, transfected into C6 cells, and mRNA levels were measured in control and amino acid-depleted cells (Fig. 1, B–F). It was previously shown that transcription of the wild-type tetcat-1 cDNA in this vector system is not affected by nutrient deprivation and can be induced by doxycycline (10). However, doxycycline was not used in these studies; the low expression under these conditions provided an ideal system for studying the accumulation of mRNA during amino acid deprivation.
The effects of amino acid deprivation on the expression of tetcat-1 and endogenous cat-1 mRNAs were examined on Northern blots, using probes (tet and 5′-cat-1) that are specific for tetcat-1 and cat-1 mRNAs, respectively. Amino acid deprivation for 4 h caused increases in both the chimeric tetcat-1 mRNA (Fig. 1B, con-wt) and the endogenous 7.9 kb cat-1 mRNA, consistent with our previous results. Similar increases in mRNA levels were seen for all the constructs that contained the 3′-terminal 217 bases. Deletions of different parts of bases 2277–6236 (tetcat-1/Δ1 – tetcat-1/Δ5 in Fig. 1, B–E) did not affect induction of the chimeric mRNAs by amino acid starvation. For all these constructs, increases of 5- to 10-fold were seen, a level similar to wild type. In the tetcat-1/Δ2 transfectant shown in Fig. 1B, the amount of cat-1/7.9 mRNA did not increase during amino acid deprivation, possibly due to titration of factors by the transfected mRNA. Consequently, samples from a second transfectant were analyzed by probing with the cat-1 probe, which hybridizes to both endogenous and tetcat-1 RNAs. In this case, amino acid deprivation stimulated the accumulation of both RNAs (Fig. 1C).
In contrast, tetcat-1/Δ6, which does not contain the 3′-terminal 758 bases (residues 5695–6453) was not induced by amino acid starvation (Fig. 1F). Two tetcat-1 mRNAs are seen. The cat-1 polyadenylation site is missing in this construct, so these two RNAs arise from polyadenylation at vector sequences. Neither of these RNAs increased during starvation. In contrast, when the same blot is probed with the cat-1 probe, which detects both endogenous and tetcat-1 RNAs, the endogenous mRNA is seen to increase. Because some of the region that is deleted in tetcat-1/Δ6 (5695–6235) is present in the tetcat-1/Δ4 and Δ5 constructs, which show the starvation-induced mRNA accumulation, we conclude that the sequences required for induction are in residues 6236–6453. These regulatory sequences are present in all of the constructs except tetcat-1/Δ6. In all cell lines, accumulation of the endogenous cat-1 and asparagine synthase (AS) mRNAs was induced by amino acid starvation, confirming the adaptive response of the cells (11).
AU-rich elements in the 3′-UTR are known to regulate mRNA stability (13). Several of these elements are found in the 759-base region that is deleted from tetcat-1/Δ6, the mRNA whose level is not increased by amino acid deprivation. There are two single AUUUA elements (position 5831 and 5858) and one overlapping AUUUUAUUUUA element (position 6268). The single AREs are probably not important for regulating stability because one is deleted in tetcat-1/Δ4 and other is deleted in tetcat-1/Δ5. Despite these deletions, both of these mRNAs accumulate to levels similar to the control during amino acid deprivation (Fig. 1E). However, deletion of the entire distal 759 bases that contains the overlapping AU motif at position 6268 resulted in complete loss of regulation of the tetcat-1/Δ6 RNA (Fig. 1F), suggesting that this sequence is important in regulation of mRNA stability. The importance of this sequence was tested directly by mutating the overlapping AU motif in the tetcat-1/Δ4 RNA (Fig. 1A). The mutated mRNA (tetcat-1/Δ4-AUmut) only showed a 50% increase during amino acid starvation (Fig. 1G), demonstrating that this 11-residue element is essential for induction of cat-1 mRNA levels. This AU-rich element is the shortest sequence known to be required for the regulation of mRNA stability (13). Furthermore, this is the first demonstration of an AU-rich sequence that regulates mRNA stability in response to amino acid starvation. The distal 217 base region of the cat-1 mRNA 3′-UTR that contains the critical 11-residue sequence will therefore be called the nutrient-sensor AU-rich element (NS-ARE).
To prove that the NS-ARE mediates the increased mRNA stability during amino acid starvation, we showed that this element functions when it is present at the 3′-end of a chimeric mRNA. To accomplish this, mRNAs from two constructs that encode chloramphenicol acetyltransferase were studied. pCAT encodes an RNA containing the SV40 late polyadenylation signal (Fig. 2A). pCAT/UTR contains nucleotides 5696–6453 from the 3′-end of the cat-1 cDNA inserted in front of the SV40 polyadenylation signal. The RNA transcript from this construct, which contains two polyadenylation signals, has the potential to give rise to two mRNAs (Fig. 2A). The amount of RNA from pCAT did not increase when transfected cells were starved for amino acids, even though there was a large increase in the amount of endogenous 7.9 kb cat-1 mRNA (Fig. 2B). In cells transfected with pCAT/UTR, two RNAs were expressed (Fig. 2C). During amino acid deprivation, there was a 3-fold increase in the amount of the smaller mRNA, which results from polyadenylation at the cat-1 polyadenylation site. This demonstrates that the NS-ARE is capable of enhancing mRNA stability in a heterologous mRNA. The amount of the larger mRNA, which results from polyadenylation at the SV40 site, also increased during deprivation, but only by 1.5-fold. This suggests that the NS-ARE stabilizes mRNAs most effectively during amino acid deprivation when it is close to the 3′-end of the molecule. In this experiment, the level of endogenous cat-1 mRNA increased by 6-fold during starvation. Because this increase is greater than those seen for the CAT/UTR RNAs, it is possible that the 757 bp from the cat-1 mRNA that are present in CAT/UTR do not contain all the sequences required for mRNA stabilization.
Fig. 2. The NS-ARE increases the stability of a chimeric mRNA during amino acid starvation.
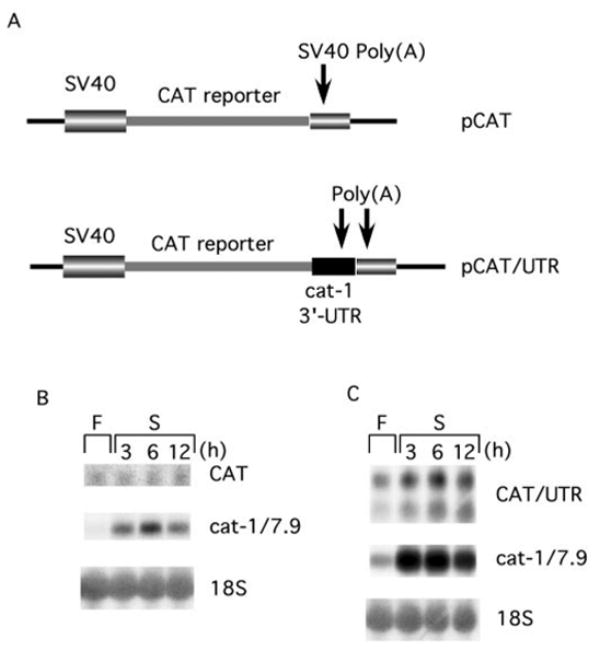
A, map of the transcripts from the pCAT and pCAT/UTR vectors. pCAT/UTR contains the 759 3′-terminal residues from the cat-1 cDNA inserted between the CAT coding sequence and the SV40 late polyadenylation region. B, cells stably transfected with pCAT were incubated in amino acid-containing (F) or -deficient (S) media for the indicated times. RNA was isolated and analyzed on Northern blots. The CAT mRNA was detected with the chloramphenicol acetyltransferase probe and the cat-1/7.9 mRNA was detected with the 5′-cat-1 probe. C, cells stably transfected with pCAT/UTR were incubated in amino acid-containing (F) or -deficient (S) media and RNAs were analyzed on Northern blots as in B.
The importance of the cat-1 NS-ARE was further supported by the sequence analysis of the mouse 3′-UTR of the cat-1 mRNA. A mouse EST clone (GenBank™ accession number, AV271225) was compared with the distal 3′-UTR of the rat cat-1 mRNA. As shown in Fig. 5A, the AUUUUAUUUUA element and the flanking sequences are fully conserved between the mouse and the rat cDNAs. This was further confirmed by sequencing of a mouse genomic DNA clone that was isolated in our laboratory (not shown).2
Fig. 5. The 11-residue AU-rich element in the NS-ARE binds cytoplasmic HuR during amino acid starvation.
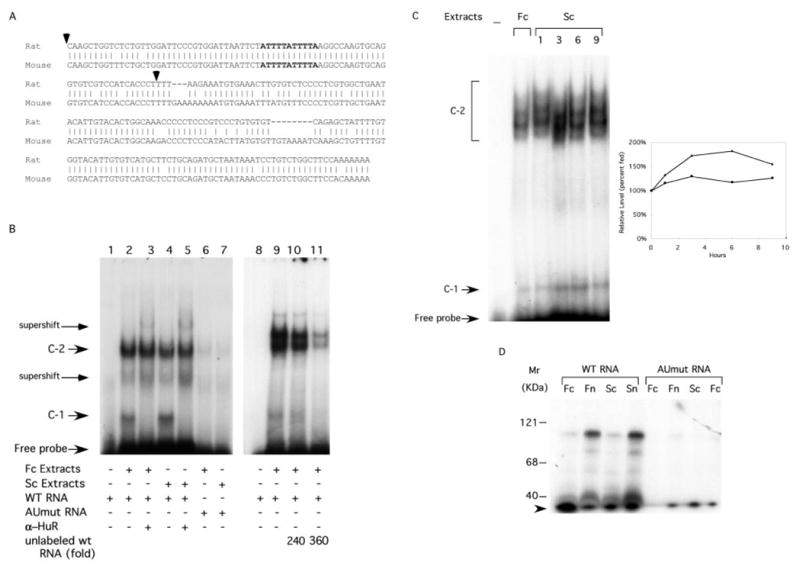
A, alignment of nucleotide sequences of the 3′-terminal end of the rat cat-1 cDNA (residues 6232–6472, Ref. 12) and a mouse cat-1 cDNA (EST AV271225). The 11-residue AU-rich element (bold) is conserved between the mouse and the rat. The arrowheads denote the sequence of the RNA used for EMSA experiments. B, EMSA analysis using cytoplasmic extracts from amino acid-fed cells (Fc) or cells deprived of amino acids for 6 h (Sc) cells and 32P-labeled RNAs. Antibodies to HuR were added to the indicated reactions. Unlabeled wild type RNA was added in the indicated molar excess over 32P-RNA. Regions of the gel are indicated (Supershift) where intensity increases when anti-HuR antibody is added. Data from one of three repetitions of the experiments are shown. C, EMSA experiment performed with wild type 32P-RNA and cytoplasmic extracts from fed cells (Fc) or cells deprived of amino acids for the indicated times (Sc). The graph shows the relative amounts of the C-1 (▲) and C-2 (●) species measured by scanning the autoradiograms. Results are means from two experiments. D, protein-RNA cross-linking. Extracts from fed cells and cells starved for 6 h were incubated with 32P-labeled wild type and mutant RNAs and cross-linked using UV irradiation. The samples were subjected to RNase digestion and analyzed by electrophoresis on SDS gels as described under “Experimental Procedures.” The arrowhead marks degradation products of the labeled RNA that runs at the dye front of the gel.
Amino Acid Starvation Induces Transient Accumulation of HuR in the Cytoplasm
HuR, is an RNA-binding protein that has been suggested to bind AREs, leading to mRNA stabilization (13, 14). The most conserved ARE sequence among all HuR-binding mRNAs is the 11 nucleotide element, AUUUUAUUUUA (14), that is required for the starvation-induced stabilization of the cat-1 mRNA. HuR contains sequences that allow it to shuttle between the nucleus and cytoplasm (15). HuR may bind target mRNAs in the nucleus and follow them to the cytoplasm, thus protecting them from degradation (19). Alternatively, increased cytoplasmic HuR levels may cause increased stability of ARE-containing mRNAs by binding in the cytoplasm (17–19).
Based on this information, we carried out experiments to determine whether HuR could be involved in the stabilization of the cat-1 mRNA during amino acid deprivation by the NS-ARE. First, we used cell fractionation and immunocytochemistry to test whether amino acid deprivation increases cytoplasmic levels of HuR. Analysis of HuR levels by Western blotting showed that amino acid starvation caused a 60% decrease of total HuR levels by 12 h (Fig. 3A). At the same time the levels of AUF1 (also known as HnRNP D), an ARE-binding protein, which is associated with increased mRNA decay (23), did not change (Fig. 3A). The specificity of the anti-AUF1 antibody was verified by performing a Western blot in the presence of the antigenic peptide p37AUF1 (Fig. 3A).
Fig. 3. Amino acid starvation induces the transient cytoplasmic accumulation of HuR.
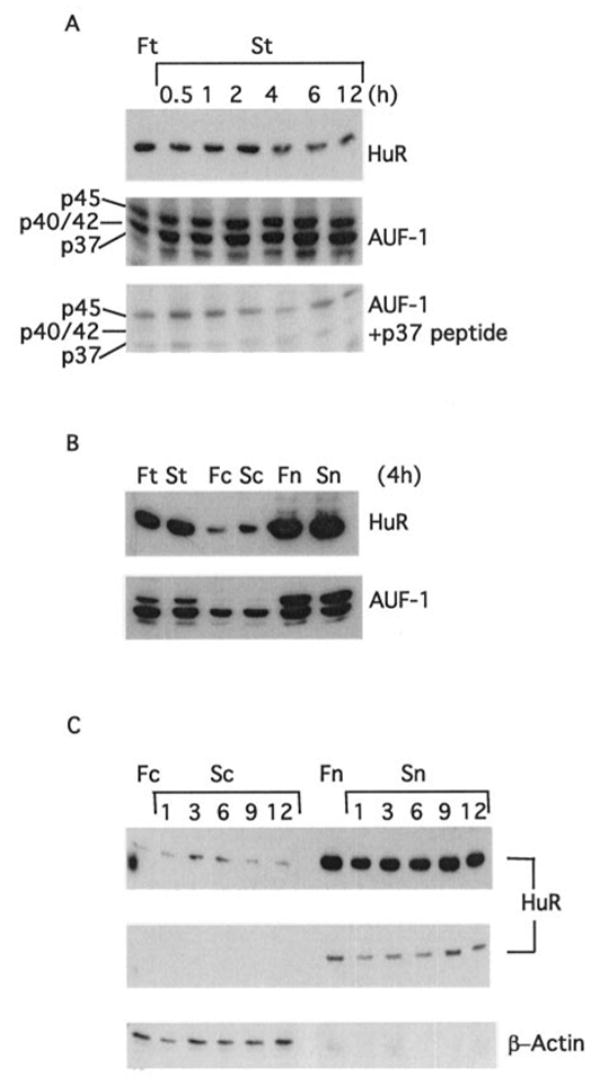
A, total cell lysates (15 μg of protein) from cells maintained in either amino acid-deficient (S) or amino acid-containing (F) medium were analyzed by immunoblotting using anti-HuR, anti-AUF1, or anti-AUF1 in the presence of 0.1 μg/ml p37 antigenic peptide. B and C, total, nuclear, and cytoplasmic extracts from C6 cells treated for the times indicated in either amino acid-containing (F) or amino acid-depleted media (S) were analyzed by immunoblotting using anti-HuR and anti-β-actin antibodies. Fc and Sc denote cytoplasmic extracts from fed and starved cells, respectively. Fn and Sn denote nuclear extracts from fed and starved cells. Two different exposures of the anti-HuR immunoblot are shown in C.
We next tested the cytoplasmic and nuclear localization of HuR, by performing Western blot analysis of cytoplasmic and nuclear fractions. In both fed and starved cells, the majority of the HuR was nuclear (Fig. 3B). However, there was a large increase in the amount of cytoplasmic HuR after 4 h of amino acid deprivation. This change was not seen for AUF1. Analysis of HuR levels during a time course of amino acid deprivation showed that cytoplasmic HuR levels increased transiently. Levels at 3 and 6 h of starvation were 5-fold higher than in fed cells and returned to control levels by 9 h (Fig. 3C). As expected, β-actin was only present in cytoplasmic extracts, confirming the purity of the cell fractions (Fig. 3C).
The findings that amino acid starvation alters HuR level and distribution are supported by immunocytochemistry (Fig. 4). In control cells HuR was largely in the nucleus. However, after 4 h of amino acid starvation, nuclear HuR levels decreased and cytoplasmic HuR levels increased (Fig. 4A). This increase was more evident in amino acid-depleted than in heat-shocked cells, a treatment that has been previously shown to increase cytoplasmic HuR levels (19). In fact, the effects of starvation are as large as those of actinomycin D, which also increases cytoplasmic HuR levels (Fig. 4B and Ref. 19). We next determined the distribution of HuR during a time course of amino acid starvation. Decreased nuclear HuR staining was visible by 1 h and the decrease continued for 9 h (Fig. 4B). It was next demonstrated that the redistribution of HuR caused by amino acid depletion is reversed by amino acid refeeding. This was accomplished by depriving cells of amino acids for 6 h and then incubating in complete medium for an additional 3 h. Refeeding of starved cells resulted in increased HuR levels in the nucleus (Fig. 4B, S6h/F3h), with a staining pattern similar to control cells. These data suggest that the cytoplasmic localization of HuR is subject to adaptive regulation in response to amino acid availability.
Fig. 4. The cytoplasmic localization of HuR is regulated by amino acid availability.
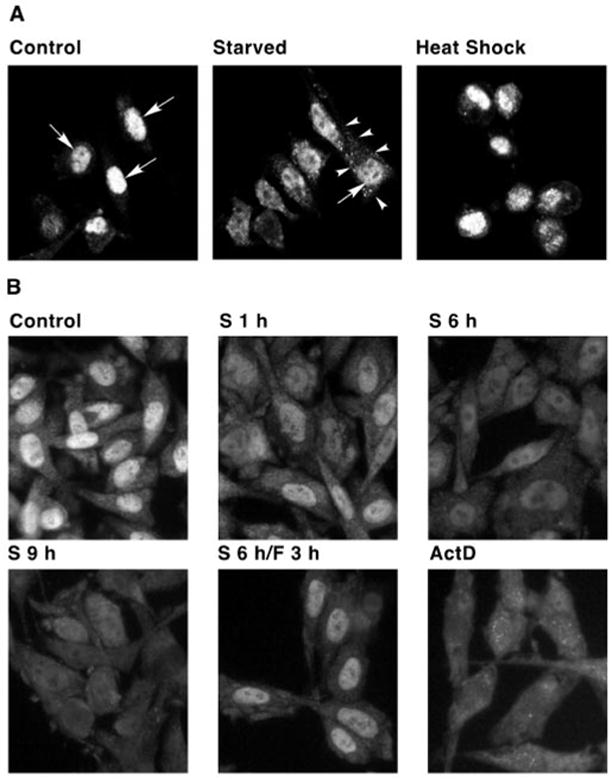
A, C6 cells were grown on chamber slides and were either incubated in amino acid-depleted medium (Starved) for 4 h or heat shocked at 45 °C for 2 h. Nuclei are marked by arrows. The margin of one starved cell is marked by arrowheads. B, cells were incubated for the indicated times in either amino acid-containing (Control) or amino acid-depleted medium (S). Cells were also treated with actinomycin D (1 μg/ml) for 4 h. One sample was starved for 6 h followed by 3 h incubation in medium containing amino acids (S6h/F3h). Cells were fixed, permeabilized, and prepared for confocal microscopy (A) or immunofluorescence microscopy (B) using an anti-HuR antibody, as described under “Experimental Procedures.”
HuR Binds to the NS-ARE of the cat-1 mRNA Via the AU-rich Element and Formation of These Complexes Increases During Amino Acid Deprivation
It is shown here (Figs. 1 and 2) that the NS-ARE is important for increased cat-1 mRNA levels during amino acid starvation. Because mRNAs are stabilized by the binding of HuR to AU-rich elements (13), it is possible that binding of HuR to the NS-ARE increases during amino acid starvation. To test this hypothesis, we studied the ability of the NS-ARE to bind HuR in vitro using electrophoretic mobility shift assays. These studies used 32P-RNA probes containing 77 nucleotides from the 3′-end of the cat-1 mRNA that contains the NS-ARE (Fig. 5A). The probes were incubated with cytoplasmic extracts and complexes were analyzed by electrophoresis on native polyacrylamide gels either directly or after addition of anti-HuR antibody. Incubation of the RNA with cytoplasmic extracts from fed cells resulted in the appearance of two intense bands, C-1 and C-2, and several fainter bands (Fig. 5B, lane 2). These shifted bands represent RNA-protein complexes. Addition of anti-HuR antibody caused the disappearance of the C-1 complex, indicating that it contains HuR (Fig. 5B, lane 3). Two species either appeared or increased in intensity when anti-HuR antibody is present (Fig. 5B, supershift). It is likely that these are derived from the C-1 complex, although the exact nature of these bands cannot be determined because of the complexity of the pattern obtained using unfractionated cytoplasmic extracts. In contrast to the results with C-1, anti-HuR antibody only caused a 10% decrease in the amount of material migrating as the C-2 complex. This result makes it likely that this complex either does not contain HuR, or it contains HuR that is not accessible to the antibodies. When the experiment was performed with cytoplasmic extracts from cells that had been starved for 6 h, the same complexes were seen. Moreover, C-1 was supershifted by anti-HuR but C-2 was not (Fig. 5B, lanes 4 and 5).
Additional experiments tested the importance of the NS-ARE in the formation of these complexes. This was accomplished by carrying out experiments using an oligonucleotide with substitutions that eliminated the 11-residue AU-rich element (Fig. 1A). This mutation in a tetcat-1 mRNA abolishes the ability of the NS-ARE to stabilize the mRNA during amino acid deprivation (Fig. 1F). EMSA experiments revealed that greatly reduced amounts of the C-1 and C-2 complexes were formed by the mutant RNA and complexes were not seen with extracts from starved or fed cells (Fig. 5B, lanes 6 and 7). The C-1 complex was not observed even in longer exposures of the autoradiogram shown in the figure (not shown). These results indicate that the AUUUUAUUUUA element is absolutely required for the formation of the complexes under all the conditions examined. Finally, we demonstrated that the complexes are specific (Fig. 5B). Formation of both the C-1 and C-2 complexes containing wild-type 32P-labeled RNA were inhibited when unlabeled wild-type competitor RNA was added to the incubations.
Our cell fractionation experiments show that there is a transient increase in cytoplasmic HuR during amino acid deprivation (Fig. 3). This finding, combined with our evidence that the C-1 complex contains HuR protein, suggests that the length of amino acid deprivation should affect the amount of the C-1 complex formed by cytoplasmic extracts in vitro. Fig. 5C shows that this is the case. Low levels of the C-1 complex were formed by extracts from fed cells. The level increased when extracts from starved cells were used. The amount increased to nearly twice that in fed cells for extracts from cells starved for 3 and 6 h and then declined. The fact that starvation caused parallel increases in the amounts of the C-1 complex and the level of cytoplasmic HuR (compare Figs. 3C and 5C) provides additional support for the importance of this protein in ribonucleoprotein complex formation on the NS-ARE. The C-2 complex is resolved into a group of bands in this experiment because the gel was run longer than the one in Fig. 5B. Amino acid starvation also caused an increase in the amount of this species, but the maximum increase was only 25%.
The EMSA experiments show that the NS-ARE can form two different complexes in vitro. Both complexes require the 11-residue AU-rich element, although C-1 contains HuR whereas C-2 probably does not. To obtain further information about the proteins that bind to the NS-ARE, we used ultraviolet light to cross-link 32P-RNA and protein. These complexes were then digested with RNase and analyzed by SDS gel electrophoresis and autoradiography (Fig 5D). Radioactively labeled proteins are ones that were cross-linked to the 32P-RNA, indicating RNA-protein association. When wild-type RNA was incubated with either nuclear or cytoplasmic extracts, labeled bands were detected. These include species at 40, 54, 75, and 100 kDa. The proteins in these labeled bands are slightly smaller than these sizes because each species contains crosslinked RNA. The 40-kDa species is the size expected for labeled HuR, consistent with results obtained by others (24, 25). It is interesting that this species is present at similar levels in nuclear and cytoplasmic extracts, even though HuR is present at much higher levels in nuclear fractions (Fig. 3B). In contrast, the 54-, 75-, and 100-kDa bands obtained from nuclear extracts were more intense than the ones from cytoplasmic extracts.
Experiments were also performed to compare the cross-linking in extracts from fed and amino acid-deprived cells. Similar results were obtained with extracts from fed and starved cells, although it appears that slightly higher levels of labeled proteins were obtained with extracts from starved cells. Finally, the importance of the NS-ARE was demonstrated by analyzing labeled proteins that were cross-linked to the mutant RNA. In this case, no labeled bands were detected. These results demonstrate the ability of the NS-ARE to form ribonucleoprotein complexes containing several proteins and the importance of the 11-residue AUUUUAUUUUA sequence in this binding.
DISCUSSION
We have shown that the cat-1 mRNA contains the NS-ARE, an element that is important in stabilizing the mRNA in response to amino acid deprivation. Characterization of deletion constructs suggests that the NS-ARE is located in the 217 nucleotides at the 3′ end of the message (Fig. 1). Moreover, the AU-rich sequence, AUUUUAUUUUA, is absolutely required for mRNA stabilization during amino acid deprivation. We do not know which other sequences in the 217-residue region are part of the NS-ARE. Further studies with chimeric mRNAs will be required to define the NS-ARE precisely.
Based on their sequences and their effects on mRNA stability, three classes of AREs have been defined (reviewed in Ref. 13). Class I AREs contain scattered copies of the AUUUA sequence within a U-rich region. Class II AREs contain overlapping AUUUA motifs within a U-rich region. Class III AREs do not contain the AUUUA motif, but they are U-rich sequences. The NS-ARE does not fit in any of the three classes. Although it contains a consensus AU-element, it lacks the U-rich regions surrounding the element. Therefore, the NS-ARE may represent a novel ARE. It will be interesting to see whether other mRNAs contain ARE sequences of this type and whether these AREs also regulate mRNA stability during amino acid deprivation.
We focused our attention on HuR because this mRNA-binding protein is known to play a role in the regulation of mRNA stability. Overexpression of HuR increases the lifetime of many ARE-containing mRNAs suggesting that HuR binding stabilizes mRNAs (15, 18, 26, 27). However, it is also possible that HuR overexpression stabilizes mRNAs by sequestering other proteins that decreases mRNA stability. An interesting feature of HuR is that it contains sequences that allow shuttling between the nucleus and the cytoplasm (15). It has been suggested that HuR binds mRNAs in the nucleus and then escorts the mRNAs to the cytoplasm where HuR protects them from degradation (19). Several cellular stresses, including UV irradiation, heat shock, and inhibition of protein synthesis increase the cytoplasmic concentration of HuR suggesting that this shift plays a role in the regulation of mRNA stability (13). In fact, it has been shown that the increased cytoplasmic concentration of HuR in UV-treated cells is well correlated with increased ARE-mediated mRNA stability (18). We have demonstrated that amino acid deprivation causes a transient increase in cytoplasmic HuR levels. Therefore this physiological cellular stress is also capable of shifting the balance of HuR between the nucleus and cytoplasm, and this shift is consistent with the proposed models of HuR function in mRNA stabilization.
We propose that the binding of HuR to the NS-ARE is important for mRNA stabilization during amino acid starvation. This argument is based on kinetic data. During amino acid deprivation, there are parallel changes in the level of cytoplasmic HuR and in the ability of cytoplasmic extracts from starved cells to form the C-1 ribonucleoprotein complex that contains HuR. Moreover, the levels of NS-ARE-containing mRNAs in vivo change during starvation with a similar time course. The chimeric tetcat-1 mRNA reaches a peak at 3–6 h of amino acid deprivation and then declines (10). Because this mRNA is expressed from a heterologous promoter that is not regulated by amino acid deprivation, the changes in level are due to changes in mRNA stability. A similar transient increase is seen for levels of the endogenous 7.9 kb cat-1 mRNA (Fig. 1F and Ref. 10). Significantly, the AS mRNA, which also increases during amino acid starvation, shows different kinetics; its level increases continuously over 24 h of starvation. Because there are parallel changes in the level of cytoplasmic HuR, in the formation of NS-ARE complexes, and in the level of NS-ARE-containing mRNA in vivo during amino acid deprivation, we propose that HuR binding plays a direct role in mRNA stabilization.
What causes the increased binding of HuR to the cat-1 mRNA in amino acid starved cells? The increased cytoplasmic HuR during amino acid starvation probably contributes to the increased binding. However, increased cat-1 mRNA stability may not be exclusively dependent on cytoplasmic HuR levels. It is possible that covalent modification of HuR or another protein in the complex could affect the specificity or affinity of one of the proteins. In fact, it has been shown that cAMP stimulates the formation of a HuR-containing complex on the SGLT1 mRNA by a mechanism that involves protein phosphorylation (24, 25, 28). In addition, it is possible that amino acid starvation causes changes in the levels of another protein that binds to the cat-1 mRNA along with HuR.
It has been speculated that HuR increases stability by protecting the mRNAs from degradation rather than by slowing down removal of the poly(A) tail (13). It was recently shown that the mammalian exosome, a 3′–5′ exonuclease involved in mRNA degradation, can bind to some AREs that also bind HuR (29, 30). These AREs enhance mRNA turnover under some conditions and stabilize the mRNA under other conditions (29). This suggests a model in which the exosome binds to the ARE and targets the mRNA for degradation. Binding of HuR to the mRNA could prevent exosome binding and stabilize the mRNA. The results of our study are consistent with such a complex mechanism. Two distinct protein complexes, C-1 and C-2 form on the NS-ARE and formation of these complexes is absolutely dependent on the 11-residue AU-rich element. In addition, at least 4 proteins were cross-linked to the NS-ARE. One of these proteins is HuR, based on its size and its ability to bind to the NS-ARE (25). The identity of the other proteins is unknown. The nature of these proteins and their interaction with the cat-1 mRNA are currently under investigation. It will be interesting to determine whether both HuR and the exosome participate in the regulation of cat-1 mRNA degradation.
It is concluded that the nutritional status of cells influences the subcellular localization and binding ability of RNA-binding proteins, such as HuR. The finding of this report is that the adaptive regulation of regulatory RNA-binding proteins is part of the response of cells to nutritional stress. The interaction of the NS-ARE in the cat-1 mRNA with HuR appears to play an important role in regulation of mRNA stability during amino acid starvation. These studies will have a significant impact on understanding the regulatory mechanisms of ARE-mediated mRNA decay by nutrient availability.
Acknowledgments
We thank Irene Krukovets and Linyin Zhou for excellent technical assistance.
The abbreviations used are
- cat-1
cationic amino acid transporter gene
- UTR
untranslated region
- AS
asparagine synthase
- NS-ARE
nutrient sensor AU-rich element
- EMSA
electrophoretic mobility shift assay
Footnotes
This work was supported by National Institutes of Health Grant R01 DK53307-01 (to M. H.) and Graduate Student Fellowship 5T32 DK07319 (to J. F.).
The sequence ATTTATTTTA (GenBank™ accession number U704476, position +6268) should read ATTTTATTTTA.
References
- 1.Pain VM. Biochimie (Paris) 1994;76:718–728. doi: 10.1016/0300-9084(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 2.Fafournoux P, Bruhat A, Jousse C. Biochem J. 2000;351:1–12. doi: 10.1042/0264-6021:3510001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruhat A, Fafournoux P. Curr Opin Clin Nutr Metab Care. 2001;4:439–443. doi: 10.1097/00075197-200109000-00016. [DOI] [PubMed] [Google Scholar]
- 4.Palacin M, Estevez R, Bertran J, Zorzano A. Physiol Rev. 1998;78:969–1054. doi: 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- 5.Closs EI. Curr Opin Nephrol Hypertens. 2002;11:99–107. doi: 10.1097/00041552-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez JM, Yaman I, Merrick WC, Koromilas AE, Wek RC, Sood R, Hensold JO, Hatzoglou M. J Biol Chem. 2001;29:29. doi: 10.1074/jbc.M109199200. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez JM, Yaman I, Sarnow P, Snider MD, Hatzoglou M. J Biol Chem. 2002;4:4. doi: 10.1074/jbc.M201052200. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez J, Bode B, Koromilas A, Diehl JA, Krukovets I, Snider MD, Hatzoglou M. J Biol Chem. 2002;277:11780–11787. doi: 10.1074/jbc.M110778200. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez J, Yaman I, Mishra R, Merrick WC, Snider MD, Lamers WH, Hatzoglou M. J Biol Chem. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
- 10.Aulak KS, Mishra R, Zhou L, Hyatt SL, de Jonge W, Lamers W, Snider M, Hatzoglou M. J Biol Chem. 1999;274:30424–30432. doi: 10.1074/jbc.274.43.30424. [DOI] [PubMed] [Google Scholar]
- 11.Hyatt SL, Aulak KS, Malandro M, Kilberg MS, Hatzoglou M. J Biol Chem. 1997;272:19951–19957. doi: 10.1074/jbc.272.32.19951. [DOI] [PubMed] [Google Scholar]
- 12.Aulak KS, Liu J, Wu J, Hyatt SL, Puppi M, Henning SJ, Hatzoglou M. J Biol Chem. 1996;271:29799–29806. doi: 10.1074/jbc.271.47.29799. [DOI] [PubMed] [Google Scholar]
- 13.Brennan CM, Steitz JA. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keene JD. Proc Natl Acad Sci U S A. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan XC, Steitz JA. Proc Natl Acad Sci U S A. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan XC, Steitz JA. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng SS, Chen CY, Xu N, Shyu AB. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. Proc Natl Acad Sci U S A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutson RG, Kilberg MS. Biochem J. 1994;304:745–750. doi: 10.1042/bj3040745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz RA, Erlanger BF, Guntaka RV. Biochim Biophys Acta. 1983;739:258–264. doi: 10.1016/0167-4781(83)90099-4. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Pascual F, Hausding M, Ihrig-Biedert I, Furneaux H, Levy AP, Forstermann U, Kleinert H. J Biol Chem. 2000;275:26040–26049. doi: 10.1074/jbc.M910460199. [DOI] [PubMed] [Google Scholar]
- 23.Laroia G, Cuesta R, Brewer G, Schneider RJ. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 24.Lee WY, Loflin P, Clancey CJ, Peng H, Lever JE. J Biol Chem. 2000;275:33998–34008. doi: 10.1074/jbc.M005040200. [DOI] [PubMed] [Google Scholar]
- 25.Loflin P, Lever JE. FEBS Lett. 2001;509:267–271. doi: 10.1016/s0014-5793(01)03176-3. [DOI] [PubMed] [Google Scholar]
- 26.Levy NS, Chung S, Furneaux H, Levy AP. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Campbell LE, Miller CM, Proud CG. Biochem J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loflin P, Lever JE. FEBS Lett. 2001;492:233–237. doi: 10.1016/s0014-5793(01)02260-8. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Kiledjian M. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]


