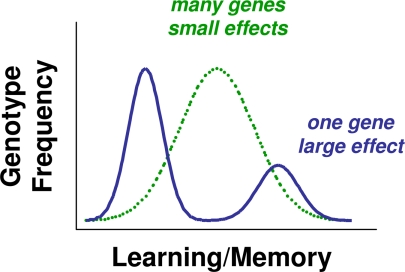
Learning, like any phenotypic trait, is expected to evolve under natural selection. A prerequisite for evolutionary change is heritable variation in learning in natural populations. It is customary in evolutionary biology to think of genetic variation in a complex trait such as learning ability in terms of a so-called “bell curve” or normal distribution (1) (Fig. 1). Most individuals are average at learning, although some may be very good and some very bad. Such a pattern of variation, if it has a genetic basis, would involve the action of multiple, probably many, genes each making a small contribution to variation in learning ability. Consistent with this view, decades of research on learning mutants of the fruit fly Drosophila melanogaster have characterized a large number of genetic loci involved in learning and memory (2, 3). Likewise, results of artificial selection on lines derived from natural populations of Drosophila (4) as well as quantitative genetic studies of other organisms [e.g., honey bees (5)] are all suggestive of a pattern of continuous genetic variation in learning and memory. In a recent issue of PNAS, work reported by Mery et al. (6) suggests that learning and memory in D. melanogaster in nature may be distributed quite differently, specifically as distinct types defined primarily by allelic variants of a single gene (Fig. 1). Their findings provide the first clear-cut evidence in non-human animals of a naturally occurring genetic polymorphism in learning and memory.
Fig. 1.
“Bell curve” of the type generally presumed to describe a pattern of genotypic variation in learning and memory in natural populations, over which is superimposed the polymorphic pattern of genetic variation that appears to be observed in D. melanogaster populations.
In documenting the learning polymorphism, Mery et al. (6) took advantage of a well established, naturally occurring genetic polymorphism at the foraging (for) locus in D. melanogaster. The for gene has been studied by Sokolowski and colleagues (7) for almost three decades and has long been associated with behavioral differences. The gene is characterized by two alleles, forR, which is dominant and which specifies a “rover” phenotype, and fors, which specifies a “sitter” phenotype. The terms “rover” and “sitter” refer to the pattern of food foraging behavior by larvae, with rover larvae moving more within and between food patches than do sitters (7, 8). In nature, the frequencies of the two phenotypes are ≈70% rovers and ≈30% sitters (7, 9).
The for gene encodes a cGMP-dependent protein kinase (PKG) (10), with rovers expressing higher levels of PKG. PKG affects a large suite of cell processes in both vertebrates and invertebrates (11–13) and has many putative functions including, in vertebrates, a possible role in learning and memory (11). Mery et al. (6) have shown that the rover–sitter polymorphism in PKG expression is indeed associated with differences in associative learning. Rover adults trained to avoid an odor in association with a mechanical shock exhibited a short-term performance in learning assays that was superior to that of sitters. This difference in short-term learning persisted even when fors was placed against a rover genetic background, isolating the for gene as the cause of these differences. Further evidence of this gene's role in learning was obtained at the level of the fly's brain. Immunostaining indicated that the for gene was strongly expressed in Drosophila mushroom bodies, regions of the brain that have been shown to be important in olfactory learning in Drosophila (2, 14). Finally, to determine whether PKG activity in the mushroom bodies might underlie differences in learning and memory between rover and sitter flies, PKG expression was experimentally up-regulated in the mushroom bodies of sitter flies. As predicted, the manipulated flies showed enhanced short-term performance in the odor-avoidance learning task.
The for gene is a candidate gene for the genetic dissection of foraging behavior, with orthologs occurring in other species, such as nematodes and honey bees, that have similar functions (15, 16). It is thus not unreasonable to expect that these genes and their PKG products could play similar roles in learning and memory in animals other than Drosophila, perhaps even humans. Does this mean that we should initiate a quest to enhance production of PKG in humans in the interest of promoting our own learning performance? Based on a second, particularly intriguing result of the behavioral assays of Mery et al. (6), the answer is “perhaps not.” Despite the reduced short-term learning in sitter flies, Mery et al. found that these flies expressed a greater, not lesser, avoidance of the conditioned odor 24 h after spaced training, reflective of superior long-term memory (LTM). In other words, rover genotypes learned faster but forgot sooner. As with the difference in short-term learning, the difference in LTM is linked to differences in mushroom body PKG activity.
Mery et al. (6) argue that the remarkable inverse relationship between short-term learning, on one hand, and LTM, on the other, may be an evolutionary consequence of the different lifestyles of rovers and sitters. Rover adults, being on the move more, may be more likely to encounter new environments (say, a different species of fruit) and thus might benefit by learning rapidly how to behave in these new environments (17). The faster rovers learn, the more beneficial that learning will be before rovers shift to yet another environment and have to learn anew.
Sitters, which are likely to remain in the same environment for a long time, would benefit particularly strongly from a highly durable LTM (18). But why, one might ask, is the LTM of rovers not equally durable? Would rovers not also benefit by having as stable a memory as possible? The answer is likely to be “no.” Holding on to obsolete memories may be costly in a number of ways. First, foraging theory suggests that forgetting may be beneficial for individuals such as rovers, which experience rapid environmental change (19). Recall of an outdated memory in a novel environment may elicit behavior that is mismatched to current conditions, which may result in reduced fitness. Moreover, old memories may interfere with the formation and recall of new, more relevant ones (20, 21). Finally, memory may incur “maintenance costs” in terms of neural tissue that ought not to be borne once stored information becomes obsolete (22). In other words, some degree of forgetting is likely to be adaptive in an ever-changing environment and so rovers, because they are presumed to change environments more frequently, should forget more readily than sitters.
The question could also be asked, might sitters not also benefit by short-term learning and memory that is as potent as that of rovers? Here, the answer would seem to be “yes.” So long as sitters benefit by learning at all, it would be best to learn maximally rapidly. The fact that sitters showed poorer short-term learning than rovers suggests that there may be an intrinsic physiological tradeoff associated with short-term learning and LTM. Tradeoffs in learning or memory may not be uncommon; in recent work on rat hippocampus, inhibition of long-term memory via suppression of neurogenesis was shown to improve some measures of working memory (23). We suggest that physiologically based learning/memory tradeoffs will be important to consider when making inferences about the evolution of learning.
One of the most exciting future directions in which the Drosophila work might be taken relates to the evolutionary forces that maintain the for polymorphism in learning and memory. This will not be an easy direction to take because an understanding of the factors that maintain the for polymorphism in learning and memory is complicated by the manifold effects of the for locus on fly behavior. In the years since the rover–sitter differences in larval movement were first described, numerous other attributes, including adult traits, have been shown to vary at this locus (Table 1). Rover larvae pupate off fruit and in the soil, whereas sitters tend to pupate on fruit (24). Adult rovers move farther after imbibing a drop of sucrose solution, have higher sucrose sensitivity, and habituate to sucrose more slowly than do adult sitters (25). Finally, rovers habituate more slowly in assays of the giant fiber jump-and-flight escape response (26). In the jargon of geneticists, the for gene exerts strongly pleiotropic effects on behavior and other traits.
Table 1.
Summary of the traits affected by the for polymorphism and their values for the rover and sitter phenotypes
| Phenotype | Rover forR | Sitter fors |
|---|---|---|
| Larval movement | More | Less |
| Adult movement | More | Less |
| Pupation site | On and off fruit | On fruit |
| Habituation: sucrose | Slower | Faster |
| Habituation: escape | Slower | Faster |
| Short-term learning | Stronger | Weaker |
| Long-term learning | Weaker | Stronger |
Strong pleiotropy at the for locus in Drosophila has important implications for the evolution of each trait, including learning, under directional selection. If the pattern of selection on other traits is congruent with selection on learning (in other words, if selection on all traits including learning favors the same directional change in PKG), then learning, as affected by PKG, will evolve more rapidly toward its optimum value than it would in the absence of pleiotropy. If the pattern of selection on other traits opposes selection on learning, then the outcome is more uncertain. Depending on the relative intensities of selection on the traits involved, learning, as influenced by PKG, may evolve more slowly toward its optimum, may not evolve at all, or may evolve further away from its optimum (27).
In the view of Mery et al. (6), the values of behavioral traits encoded by each for allele are coadapted in D. melanogaster. Selection apparently favors a high PKG level that defines one set of states for the various foraging traits involved while simultaneously favoring a low PKG level defining a distinctly different set of states. Intermediate PKG levels defining intermediate states in the various foraging traits, including learning and memory, are presumably disfavored by selection, resulting in a complex foraging polymorphism. In D. melanogaster, a suite of foraging traits, including learning and memory, thus appears to be evolving as a polymorphic behavioral syndrome (28).
Mery et al. (6) have opened the door for future work on ecologically relevant genetic variation in learning. First, although this work has highlighted the importance of one gene, there are hundreds of genes known to be involved in learning and behavior in Drosophila (2, 3, 29, 30). Future research may consider the role of “keystone” regulatory genes with large pleiotropic effects, relative to other genes, in the evolution of learning in Drosophila and other species. Second, given the large body of laboratory work on learning in model systems such as Drosophila, the time seems ripe for studies of the relative fitnesses of alternative learning genotypes under natural conditions (31). Understanding of the true significance of genetic variation in learning on evolutionary change will benefit greatly from in situ studies of natural populations. Drosophila's small size, rapid dispersal abilities, and propensity to colonize ephemeral habitats present distinct challenges for such studies from a behavioral ecology standpoint (however, see refs. 32 and 33). A candidate gene approach (16) in which the study of genetic variation in PKG and learning is extended from Drosophila to other, more ecologically tractable organisms, such as honey bees, butterflies, parasitic wasps, and perhaps even mice and birds, may provide a fruitful avenue of future research on the evolution of learning.
Footnotes
The authors declare no conflict of interest.
See companion article on page 13051 in issue 32 of volume 104.
References
- 1.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 2.Dubnau J, Tully T. Annu Rev Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- 3.Waddell S, Quinn WG. Annu Rev Neurosci. 2001;24:1283–1309. doi: 10.1146/annurev.neuro.24.1.1283. [DOI] [PubMed] [Google Scholar]
- 4.Mery F, Kawecki TJ. Proc Natl Acad Sci USA. 2002;99:14274–14279. doi: 10.1073/pnas.222371199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra SBC, Hunt GJ, Cobey S, Smith BH. Behav Genet. 2001;31:275–285. doi: 10.1023/a:1012227308783. [DOI] [PubMed] [Google Scholar]
- 6.Mery F, Belay AT, So AK-C, Sokolowski MB, Kawecki TJ. Proc Natl Acad Sci USA. 2007;104:13051–13055. doi: 10.1073/pnas.0702923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokolowski MB. Behav Genet. 1980;10:291–302. doi: 10.1007/BF01067774. [DOI] [PubMed] [Google Scholar]
- 8.Pereira HS, Sokolowski MB. Proc Natl Acad Sci USA. 1993;90:5044–5046. doi: 10.1073/pnas.90.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokolowski MB, Pereira HS, Hughes K. Proc Natl Acad Sci USA. 1997;94:7373–7377. doi: 10.1073/pnas.94.14.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, Coulthard A, Pereira HS, Greenspan RJ, Sokolowski MB. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Robinson PJ. J Neurochem. 1997;68:443–456. doi: 10.1046/j.1471-4159.1997.68020443.x. [DOI] [PubMed] [Google Scholar]
- 12.Müller U. Prog Neurobiol. 1997;51:363–381. doi: 10.1016/s0301-0082(96)00067-6. [DOI] [PubMed] [Google Scholar]
- 13.Bicker G. Cell Tissue Res. 2001;303:137–146. doi: 10.1007/s004410000321. [DOI] [PubMed] [Google Scholar]
- 14.Heisenberg M. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 15.Fitzpatrick MJ, Sokolowski MB. Integr Comp Biol. 2004;44:28–36. doi: 10.1093/icb/44.1.28. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick MJ, Ben-Shahar Y, Smid HM, Vet LEM, Robinson GE, Sokolowski MB. Trends Ecol Evol. 2005;20:96–104. doi: 10.1016/j.tree.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 17.Dukas R. In: Cognitive Ecology: The Evolutionary Ecology of Information Processing and Decision Making. Dukas R, editor. Chicago: Univ of Chicago Press; 1998. pp. 129–174. [Google Scholar]
- 18.Stamps J. Am Nat. 1995;146:41–58. [Google Scholar]
- 19.McNamara JM, Houston AI. J Theor Biol. 1987;125:385–395. doi: 10.1016/s0022-5193(87)80209-6. [DOI] [PubMed] [Google Scholar]
- 20.Dukas R. Anim Behav. 1995;49:1481–1490. [Google Scholar]
- 21.Cheng K, Wignall AE. Anim Cognit. 2006;9:141–150. doi: 10.1007/s10071-005-0012-5. [DOI] [PubMed] [Google Scholar]
- 22.Dukas R. J Theor Biol. 1999;197:41–50. doi: 10.1006/jtbi.1998.0856. [DOI] [PubMed] [Google Scholar]
- 23.Saxe MD, Malleret G, Vronskaya S, Mendez I, Garcia AD, Sofroniew MV, Kandel ER, Hen R. Proc Natl Acad Sci USA. 2007;104:4642–4646. doi: 10.1073/pnas.0611718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokolowski MB. J Insect Physiol. 1985;31:857–864. [Google Scholar]
- 25.Scheiner R, Sokolowski MB, Erber J. Learn Mem. 2004;11:303–311. doi: 10.1101/lm.71604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engel JE, Xie X-J, Sokolowski MB, Wu C-F. Learn Mem. 2000;7:341–352. doi: 10.1101/lm.31600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen TF. BioEssays. 2003;69:83–94. [Google Scholar]
- 28.Sih A, Bell AM, Johnson JC, Ziemba RE. Q Rev Biol. 2004;79:241–277. doi: 10.1086/422893. [DOI] [PubMed] [Google Scholar]
- 29.Greenspan RJ. Annu Rev Neurosci. 2004;27:79–105. doi: 10.1146/annurev.neuro.27.070203.144323. [DOI] [PubMed] [Google Scholar]
- 30.Kendler KS, Greenspan RJ. Am J Psychiatry. 2006;163:1683–1694. doi: 10.1176/ajp.2006.163.10.1683. [DOI] [PubMed] [Google Scholar]
- 31.Reaume CJ, Sokolowski MB. Curr Biol. 2006;16:R623–R628. doi: 10.1016/j.cub.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 32.Jaenike J. Evolution (Lawrence, Kans) 1985;39:362–369. doi: 10.1111/j.1558-5646.1985.tb05673.x. [DOI] [PubMed] [Google Scholar]
- 33.Stamps J, Buechner M, Alexander K, Davis J, Zuniga N. Anim Behav. 2005;70:609–618. [Google Scholar]