Abstract
Although concentrated soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) drive liposome fusion and lysis, the fusion of intracellular membranes also requires Rab GTPases, Rab effectors, SM proteins, and specific regulatory lipids and is accompanied by little or no lysis. To rationalize these findings, we generated yeast strains that overexpress all four vacuolar SNAREs (4SNARE++). Although vacuoles with physiological levels of Rab, Rab effector/SM complex, and SNAREs support rapid fusion without Rab- and SNARE-dependent lysis, vacuoles from 4SNARE++ strains show extensive lysis and a reduced need for the Rab Ypt7p or regulatory lipids for fusion. SNARE overexpression and the addition of pure homotypic fusion and vacuole protein sorting complex (HOPS), which bears the vacuolar SM protein, enables ypt7Δ vacuoles to fuse, allowing direct comparison of Rab-dependent and Rab-independent fusion. Because 3- to 40-fold more of each of the five components that form the SNARE/HOPS fusion complex are required for vacuoles from ypt7Δ strains to fuse at the same rate as vacuoles from wild-type strains, the apparent forward rate constant of 4SNARE/HOPS complex assembly is enhanced many thousand-fold by Ypt7p. Rabs function in normal membrane fusion by concentrating SNAREs, other proteins (e.g., SM), and key lipids at a fusion site and activating them for fusion without lysis.
Keywords: homotypic fusion and vacuole protein sorting complex, membrane fusion, Rab GTPase, yeast vacuoles
Trafficking between organelles requires selective membrane tethering, the establishment of fusion-competent microdomains, and bilayer rearrangements to allow lipid and aqueous content mixing without organelle rupture (1). Tethering and fusion microdomain assembly are governed by Rab/Ypt family GTPases and “effector” proteins that bind selectively to their GTP-bound form (2). Fusion-competent microdomains become enriched in selected lipids and proteins such as Rabs, Rab effectors, and soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (3–6). SNARE proteins are defined by their heptad repeat domains that can form four-helical bundles, in cis (with each SNARE anchored to the same membrane) or in trans (with SNAREs anchored to apposed membranes). Trans-SNARE complexes bring membrane bilayers into close apposition, but close apposition does not suffice for fusion (7). Fusion requires dramatic lipid rearrangements, which may be facilitated by local concentrations of bilayer-disrupting lipids (3, 8). Trans-SNARE bundles may promote fusion by conferring physical strain on the bilayers (9), through their bilayer-disrupting transmembrane domains (10), and by regulating the enrichment of lipids such as diacylglycerol and phosphoinositides at fusion sites (3, 4).
Rearrangement of bilayer lipids is inherent to both membrane rupture (lysis) and membrane fusion. Model liposome studies have shown that agents that promote fusion, such as PEG, also promote lysis (7), and even the fusion of cells is accompanied by some lysis (11). SNAREs promote lipid mixing between liposomes (12) and mixing of their luminal contents (13) but this mixing is accompanied by substantial lysis followed by bilayer reannealing (14, 15). Low levels of SNAREs with synaptotagmin and Ca2+ reconstitute fusion with minimal lysis (16), yet most intracellular fusion events involve a Rab family GTPase, Rab effector, and SM protein, not a synaptotagmin family member or Ca2+. The relative amounts of SNARE-driven lysis and fusion in liposomes are unknown, and the relationship between lysis and fusion in intact organelles after Rab-dependent and Rab-effector-dependent docking is unclear.
Vacuoles (lysosomes) of Saccharomyces cerevisiae provide a facile model of membrane fusion. Vacuole fusion is studied by in vitro assays (17, 18), which measure the mixing of vacuole luminal compartments. Vacuole tethering requires the Ypt7p Rab family GTPase (19) and homotypic fusion and vacuole protein sorting complex (HOPS) (20), a hexameric complex of Vps11p, Vps16p, Vps18p, Vps33p, Vps39p, and Vps41p. Vacuole fusion is regulated by the phosphorylation state of HOPS (21), which has direct affinities for Ypt7p (22), phosphoinositides (20), and SNAREs (20, 23, 24). Vacuole fusion requires four SNAREs: the Q-SNAREs Vam3p, Vam7p, and Vti1p and the R-SNARE Nyv1p. Vam7p has no hydrophobic membrane anchor, but has an N-terminal Phox homology domain (PX) that promotes its membrane association (25). Sec18p (N-ethylmaleimide-sensitive factor) and Sec17p (α-SNAP) disassemble cis-SNARE complexes, freeing them for association in trans during docking. Each vacuole in a tethered pair is drawn against the other until each has a disk-like domain of closely apposed membrane, termed the “boundary membrane” (5). Proteins (Ypt7p, HOPS, and SNAREs) and lipids (ergosterol, phosphoinositides, and diacylglycerol) that are required for fusion show interdependent enrichment at a ring-shaped microdomain, the “vertex ring,” surrounding these apposed boundary membrane discs (3, 6). Fusion proceeds around the vertex ring, joining the apposed boundary membranes to form a luminal vesicle within the larger, fused organelle. Although physiological vacuole rupture has not been observed, the cytosol contains low Mr peptide inhibitors of vacuole luminal proteases, suggesting that rupture occasionally occurs (26). During in vitro incubation, vacuoles release luminal calcium (27, 28) and become inactive for fusion (29), although it is not known whether rupture causes this calcium release or fusion inactivation.
Do SNAREs alone drive fusion, lysis, or both? If SNAREs alone can drive fusion, what are the roles of Rabs, Rab effectors, and SM proteins? Without their Rab, how efficient are SNAREs as a fusion engine? To address these questions and compare the fusion properties of organelles with those of liposomes bearing pure SNAREs (without synaptotagmin), we have introduced an assay of lysis to our vacuole fusion tester strains and genetically and biochemically regulated the levels of SNAREs, the Ypt7p Rab, and HOPS on these vacuoles. Our findings reproduce the Rab-independent fusion seen with liposomes bearing high levels of SNAREs, show that this fusion is accompanied by far more lysis than is seen with wild-type vacuoles, and provide a direct comparison of the efficiency of the Rab-dependent and Rab-independent pathways. These studies support the unifying concept that trans-SNARE pairs facilitate bilayer rearrangements and reconcile recent findings that very high levels of SNAREs alone can induce liposomes to either rupture or fuse. Our working model is that lysis is driven by a higher concentration of SNAREs at the vertex ring microdomain than is needed for fusion, and that Rab, Rab effector, and regulatory lipids are essential for efficient fusion with minimal lysis.
Results
We have modified the in vitro vacuole fusion assay to permit simultaneous assay of lysis. Vacuoles are isolated from two strains (Fig. 1A). One has normal proteases but is deleted for PHO8, encoding the major vacuolar phosphatase. The other strain has a wild-type PHO8 gene but deletions in vacuolar protease genes; without proteases, vacuoles accumulate catalytically inactive pro-Pho8p. Although neither vacuole population has active phosphatase, fusion allows proteases to gain access to pro-Pho8p and cleave it to active Pho8p, which is assayed colorimetrically. This in vitro assay is performed in the continuous presence of excess I2B, a potent cytoplasmic inhibitor of the major vacuole protease, so that pro-Pho8p is not activated via vacuole lysis. To allow a concurrent assay of lysis, we have genetically targeted GFP to the vacuole lumen. In vitro incubations with these vacuoles can be assayed for fusion by Pho8p activity and for lysis by measuring the nonsedimentable GFP.
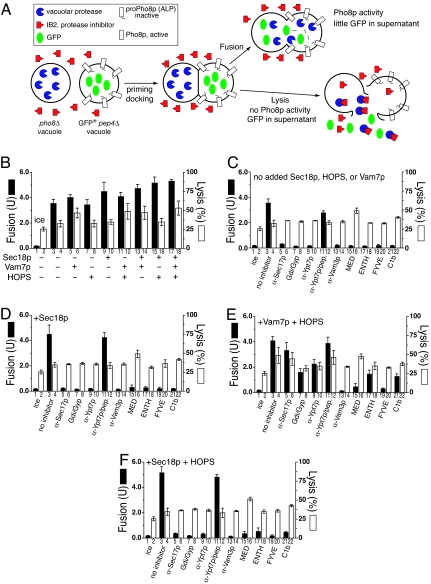
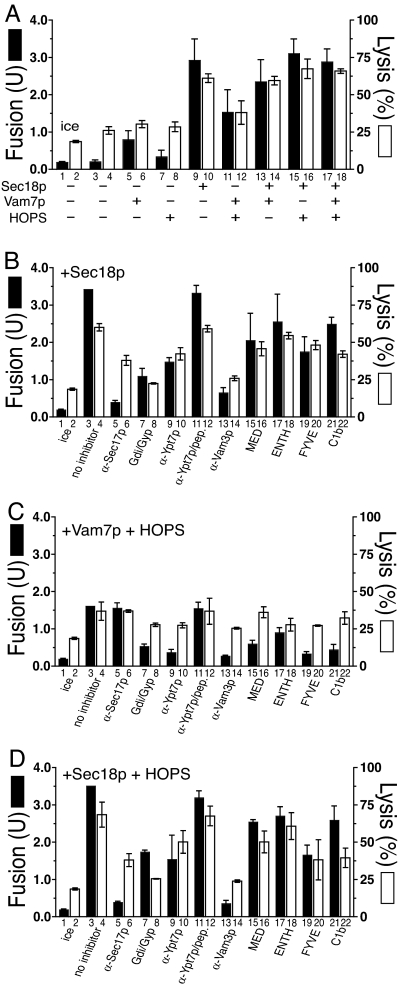
Fig. 1.
The soluble vacuolar SNARE Vam7p promotes lysis during vacuole fusion. (A) Assay schematic; see Results for details. Vacuoles from BJ3505 GFP+ and DKY6281 were used to assay vacuole fusion and lysis (see Materials and Methods). Fusion (filled bars) and percent GFP release (open bars) were measured after 60 min. (B–F) Reactions bore: no inhibitors (B), reaction pathway inhibitors without additional fusion components (C), inhibitors with rSec18p (D), inhibitors with rVam7p and HOPS (E), and inhibitors with rSec18p and HOPS (F). See Materials and Methods for all reagent concentrations. Results are the mean of three independent experiments ± SD.
Rab, HOPS, and SNAREs Drive Fusion But Not Lysis.
Vacuole fusion is temperature-dependent (Fig. 1B, bars 1 and 3). Vacuoles bear all of the proteins needed for fusion; thus, there was little stimulation by adding purified Sec18p chaperone, Vam7p SNARE, HOPS, or combinations thereof (Fig. 1B, filled bars). Without added Sec18p, Vam7p, or HOPS, fusion is blocked by antibodies to either the SNARE Vam3p or the Rab Ypt7p, by a mixture of Gyp1–46p and Gdi1p, which efficiently extracts Ypt7p (30), or by ligands (MED, ENTH, FYVE, and C1b) to key “regulatory” lipids (3) (Fig. 1C). Aliquots of these same incubations were assayed for vacuole lysis (Fig. 1C, open bars) by sedimenting the vacuoles and measuring the GFP in the supernatants. Although some lysis had occurred during vacuole isolation (Fig. 1B, bar 2), there was only a low rate of further lysis during the incubation (Fig. 1B, bar 2 vs. 4; for kinetics, see square symbols in Fig. 5B). Although the phosphoinositide ligand MED caused a modest stimulation of lysis (Fig. 1C, bar 16), none of the other inhibitory Rab, SNARE, or regulatory lipid ligands diminished this temperature-dependent lysis (Fig. 1C, open bars; see Fig. 5B, circles). Thus the efficient fusion of vacuoles bearing normal levels of Rab, Rab effectors, and SNAREs is not accompanied by detectable Rab- and SNARE-dependent lysis.
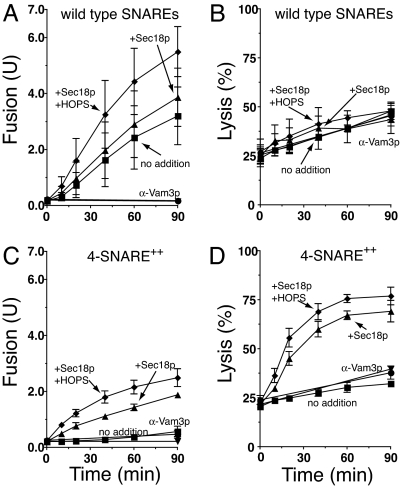
Fig. 5.
Kinetics of fusion and lysis. Standard fusion and lysis reactions (×16 scale) bore vacuoles from BJ3505 GFP and DKY6281 or 4SNARE-overproducing strains BJ3505 4SNARE++ GFP, DKY6281 4SNARE++. Two 30-μl aliquots were withdrawn before incubation at 27°C for fusion and GFP release analysis (see Materials and Methods). Additional 30-μl aliquots were withdrawn at 10, 20, 40, 60, and 90 min. Aliquots withdrawn for fusion analysis remained at 0°C for the rest of the experiment, while the separation of aliquots into pellets and supernatants was performed immediately after each sample was withdrawn. Reactions contained either: no extra additions (■), rSec18p (▴), rSec18p and α-Vam3p IgG (▾), rSec18p and HOPS (♦), or rSec18p, HOPS, and α-Vam3p IgG (○). α-Vam3p-inhibited reactions only show 0- and 90-min assays for fusion and lysis. (A and B) Fusion (A) and lysis (B) for vacuoles with physiological levels of wild-type SNARE proteins. (C and D) Fusion (C) and lysis (D) for 4SNARE++ vacuoles. α-Vam3p IgG was added to 2.7 μM for reactions containing 4SNARE++ vacuoles. Results are the mean of three independent experiments ± SD.
Excess SNARE Pairs Drive Lysis and Fusion.
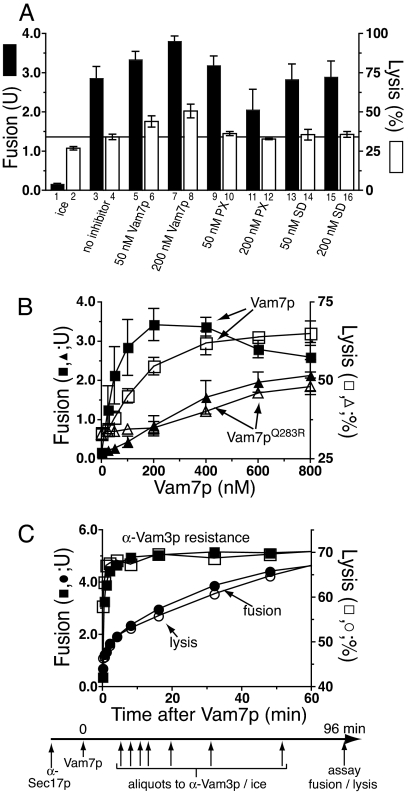
The addition of the Vam7p SNARE, which drives further trans-SNARE complex formation (31), promotes additional lysis (Fig. 1B, bar 4 vs. bar 6; P < 0.01). The full-length Vam7p is required for this lysis, as neither its PX domain nor SNARE domain will suffice (Fig. 2A). Vam7p promotes lysis through SNARE pairing, as disturbing the crucial 4SNARE 0-layer by substitution of the Vam7p Gln by Arg (Vam7pQ283R; ref. 32) caused equivalent loss of potency for fusion as for lysis (Fig. 2B). The Vam7p-triggered lysis was analyzed kinetically (Fig. 2C) in reactions that were synchronized at the step of trans-SNARE pairing (28). Vacuoles were incubated with antibody to Sec17p, preventing the release of Vam7p from cis-SNARE complexes and hence blocking trans-SNARE pairing. After 20 min, this block was bypassed by adding recombinant Vam7p (33), permitting rapid trans-SNARE pairing and consequent resistance to antibody to Vam3p. The completion of fusion, assayed by the maturation of Pho8p upon aqueous compartment mixing, takes far longer (Fig. 2C and ref. 29). Strikingly, vacuole lysis occurs in parallel with aqueous compartment mixing (Fig. 2C), suggesting that trans-SNARE pairing triggers slow lipid rearrangements that lead to either lysis or fusion. Lysis is not a simple consequence of fusion, as there is no detectable Rab- and SNARE-dependent lysis in standard fusion reactions (Fig. 1C).
Fig. 2.
Vam7p promotes lysis through SNARE pairing. (A) The full-length Vam7p is needed to promote lysis. Full-length Vam7p, its Phox homology domain (PX), or its SNARE domain (SD) was added to standard fusion and lysis assays at the indicated concentrations. Fusion (filled bars) and lysis (open bars) were measured after 60 min. (B) A 3Q:1R SNARE 0-layer is as necessary for Vam7p promotion of lysis as for fusion. Fusion reactions without ATP (33), using vacuoles from BJ3505 GFP+ and DKY6281, were examined for fusion (filled symbols) and lysis (open symbols) upon the addition of either wild-type Vam7p (squares) or Vam7pQ283R (triangles). Results in A and B are the mean of three independent experiments ± SD. (C) The kinetics of content mixing and GFP release in a synchronized vacuole fusion reaction. Fusion reactions containing 3 μg BJ3505 vacuoles bearing GFP and 3 μg DKY6281 vacuoles were incubated with affinity-purified anti-Sec17p (154 nM) for 20 min. After recombinant Vam7p (1 μM) was added, portions of the reactions (90 μl) received anti-Vam3p IgG (444 nM) or were placed on ice at indicated times (−0.5, 0.5, 1, 2, 4, 8, 16, 32, 48, 64, 80, or 96 min) and 30 μl was assayed for alkaline phosphatase activity as a measure of fusion (content mixing) after 96 min. For lysis, 30 μl was centrifuged (5,200 × g, 4°C, 6 min), and GFP fluorescence was measured in supernatants and resuspended pellets.
HOPS is a component of fusion-competent SNARE complexes (24), and the addition of both HOPS and Vam7p promotes more trans-SNARE complex formation than either alone (31). To our surprise, the addition of HOPS and Vam7p (Fig. 1E) renders fusion far more resistant to either antibody to Ypt7p (Fig. 1E, bar 9) or to the extraction of Ypt7p by the combined actions of Gdi1p/Gyp1–46 proteins (Fig. 1E, bar 7) than was seen when HOPS and Vam7p were not added (Fig. 1C) or when extra Sec18p was added, either alone (Fig. 1D) or in combination with HOPS (Fig. 1F). While these purified vacuoles bore wild-type levels of all fusion factors, supplementation with purified HOPS and Vam7p (Fig. 1E) allowed them to escape the normally strict requirement for the Ypt7p Rab, although at the price of enhanced lysis.
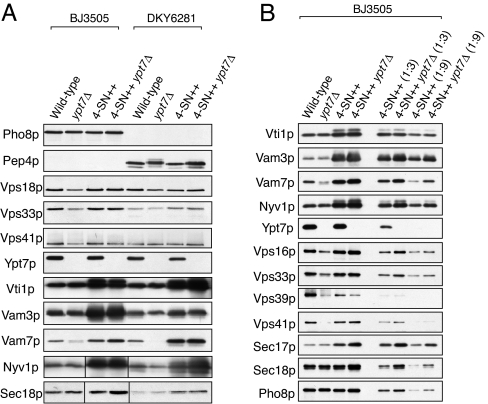
To further explore the roles of the Ypt7p Rab and vacuole SNAREs in lysis and fusion, we created 4SNARE++ strains with the expression of all four vacuolar SNAREs under constitutively strong ADH1 promoters. These were combined with YPT7 or ypt7Δ in either the protease-deficient BJ3505 background with vacuolar GFP or the Pho8p-deficient DKY6281 background. Vacuoles from each strain were analyzed by immunoblot (Fig. 3). The SNAREs were 3- to 18-fold overexpressed, recruiting substantially more Sec17p to the vacuole, whereas the levels of Sec18p, Ypt7p, or HOPS subunits were little changed by altering SNARE levels. As reported (34), almost no Vam7p was found on the vacuole in a ypt7Δ strain, but we found that the vacuole association of Vam7p was restored by the concomitant overexpression of all four SNAREs.
Fig. 3.
Immunoblot analysis of purified vacuoles. (A) Vacuoles were isolated from indicated yeast strains, and protein compositions were analyzed by immunoblot. Immunoblots for Nyv1p and Sec18p were from one gel, but lanes 1–4 were reordered for clarity. (B) Vacuoles were purified from BJ3505, BJ3505 ypt7Δ, BJ3505 4SNARE++, and BJ3505 4SNARE++ ypt7Δ, and 3 μg (lanes 1–4), 1 μg (lanes 5 and 6), or 0.33 μg (lanes 7 and 8) was analyzed by immunoblot.
Vacuoles bearing high levels of SNAREs do not fuse during in vitro incubations (Fig. 4A, bar 3), perhaps because SNARE overexpression recruits proportionately less Sec18p than Sec17p to the vacuoles (Fig. 3). The fusion of 4SNARE++ vacuoles is activated by added Vam7p and HOPS or by Sec18p (Fig. 4A, bars 9 and 11). Almost all of the Vam7p on vacuoles is present in cis-SNARE complexes (33). The stimulation of 4SNARE++ vacuole fusion by Sec18p suggests that the Sec18p bound to these vacuoles is insufficient to provide enough Vam7p to drive fusion. Sec18p-activated fusion (Fig. 4B) remains sensitive to antibody to Sec17p or Vam3p, but is inhibited far less by either α-Ypt7p, a mixture of Gyp1–46 and Gdi1p (to inactivate and extract the Rab Ypt7p), or each of the lipid ligands than is the fusion of vacuoles bearing wild-type levels of SNAREs when supplemented with Sec18p (Fig. 1D). The need for Ypt7p for fusion of 4SNARE++ vacuoles, although reduced, may not be eliminated. Strikingly, vacuole lysis is dramatically enhanced by the combination of SNARE overexpression and incubation with Sec18p (compare Fig. 4A, bar 10 vs. bars 2 and 4 with these same bars in Fig. 1D). Lysis in these incubations was reduced by Gyp1–46 plus Gdi1p (Fig. 4B, bar 8), in accord with the Ypt7p function of concentrating SNAREs at the vertex ring (6) and lysis requiring the very highest SNARE concentrations at this microdomain. Kinetic analysis of the fusion and lysis of vacuoles with wild-type or elevated levels of SNAREs (Fig. 5) confirms that normal vacuole fusion (Fig. 5A) entails little SNARE-dependent (and thus α-Vam3p-sensitive) lysis (Fig. 5B), whereas SNARE overexpression and the disassembly of these cis complexes of SNAREs by Sec18p leads to fusion (Fig. 5C) and strongly enhanced lysis (Fig. 5D), which can be blocked by α-Vam3p.
Fig. 4.
Fusion and lysis of vacuoles with high levels of SNARE proteins. Standard fusion and lysis reactions using vacuoles from BJ3505 4SNARE++ GFP and DKY6281 4SNARE++. Fusion (filled bars) and percent GFP release (open bars) were measured after 60 min: no inhibitors (A), inhibitors with rSec18p (B), inhibitors with rVam7p and HOPS (C), and inhibitors with rSec18p and HOPS (D). The fusion data in B–D were normalized to uninhibited fusion levels as follows: 3.47 ± 1.3 units (B), 1.62 ± 0.71 units (C), or 3.52 ± 0.86 units (D). Anti-Vam3p-inhibited reactions contained 2.7 μM or 5.5 μM IgG (D). Reactions with anti-Sec17p contained 366 nM IgG. Results are the mean of three independent experiments ± SD.
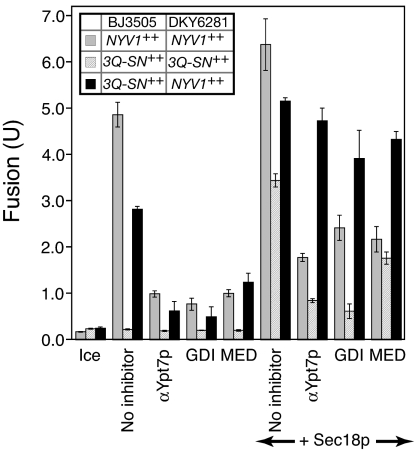
With the overexpression of each SNARE, their disassembly from cis-SNARE complexes by Sec18p, and the supply of exogenous HOPS, SNAREs may reach the required levels at the vertex ring without Ypt7p-mediated and regulatory lipid-mediated concentration, resulting in relative insensitivity of fusion to Ypt7p-targeted ligands. Only vacuole pairs with overexpressed SNAREs distributed on the two fusion partners so as to yield enhanced trans-SNARE formation show this resistance to fusion inhibitors (Fig. 6). For this study, our standard fusion test strains were prepared with selective overexpression of either all three Q-SNAREs or only the R-SNARE Nyv1p. When both fusion partners had overexpressed Nyv1p, or when they each bore enhanced levels of each of the three Q-SNAREs (and were incubated with additional Sec18p), fusion remained largely sensitive to α-Ypt7p, Gdi1p, or MED (Fig. 6). However, when one fusion partner bore overexpressed Nyv1p and the other had co-overexpression of the three Q-SNAREs, fusion was relatively insensitive to these ligands, likely because of enhanced levels of trans-SNARE complex formation.
Fig. 6.
Ypt7p Rab GTPase bypass fusion via 4SNARE overproduction requires trans-SNARE complexes. Fusion reactions (see Materials and Methods) with vacuoles isolated from BJ3505 NYV1++ and DKY6281 NYV1++ (dark gray bars), BJ3505 3Q++ and DKY6281 3Q++ (light gray bars), or BJ3505 3Q++ and DKY6281 NYV1++ (black bars) were incubated for 90 min on ice or at 27°C in the absence or presence of the indicated reagents. Results are the mean of three independent experiments ± SD.
Excess SNAREs Bypass the Ypt7p Rab.
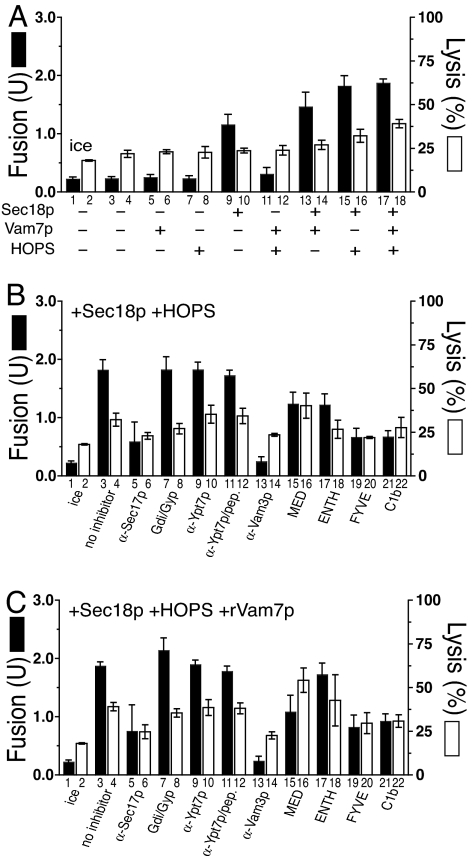
The fusion of 4SNARE++ vacuoles in the presence of added Sec18p and HOPS was only 50% inhibited by Gyp1–46 and Gdi1p (Fig. 4D). We asked whether this remaining fusion represents a component of the fusion that had bypassed the need for Ypt7p, as overexpression of the SNAREs Bet1p and Sec22p can bypass the absence of the Rab Ypt1p for ER to Golgi traffic (35, 36), or whether all of the fusion still required a reduced level of Ypt7p. Vacuoles with 4SNARE++ and ypt7Δ showed no fusion (Fig. 7A, bar 3) beyond the ice control (Fig. 7A, bar 1). Fusion was restored by added Sec18p (Fig. 7A, bar 9), especially in combination with Vam7p (Fig. 7A, bar 13), HOPS (Fig. 7A, bar 15), or both (Fig. 7A, bar 17). The fusion with HOPS and Sec18p was insensitive to antibody to Ypt7p (Fig. 7B, bar 9), as expected, and showed no inhibition (Fig. 7B, bar 7) by levels of Gdi1p and Gyp1–46p, which completely blocked the fusion of wild-type vacuoles (Fig. 1F), suggesting that this fusion represents a true Rab bypass and not merely the substitution of some other Rab. Without SNARE overexpression, the addition of HOPS, Vam7p, and Sec18p in all combinations did not restore any measurable fusion to ypt7Δ vacuoles (data not shown). Although adding Sec18p, Vam7p, and HOPS to the 4SNARE++ ypt7Δ vacuoles induced additional lysis (Fig. 7A, bar 4 vs. bar 18; P < 0.01), which was independent of Rab function (Fig. 7C, bars 4 and 8), these vacuoles showed less lysis than their YPT7 counterparts (compare Fig. 4A, bar 18 with Fig. 7A, bar 18), consistent with the concept that Ypt7p concentrates the overexpressed SNAREs at the vertex for maximal lysis. The small size of ypt7Δ or 4SNARE++ vacuoles precludes direct assay of vertex enrichment in these studies.
Fig. 7.
Fusion and lysis of ypt7Δ, 4SNARE++ vacuoles. Standard fusion and lysis reactions with vacuoles from BJ3505 4SNARE++ GFP ypt7Δ and DKY6281 4SNARE++ ypt7Δ. (A) Fusion (filled bars) and lysis (open bars) for uninhibited reactions. (B) Fusion and lysis reactions containing rSec18p and HOPS plus reaction inhibitors. (C) Reaction inhibitors in the presence of rSec18p, HOPS, and rVam7p. α-Vam3p-inhibited reactions contained 2.7 μM IgG. Results are the mean of three independent experiments ± SD.
Discussion
Membrane perturbants that promote liposome fusion also promote lysis (37, 38). However, it had been unclear whether the close relationship between lysis and fusion would also be seen for Rab, Rab effector, and SNARE-dependent fusion between biological membranes. We now show that the physiological balance of Rab, HOPS, and SNAREs drives fusion but does not promote lysis, whereas excess SNAREs drive both fusion and lysis. Excess SNARE pairing is induced by either adding extra Vam7p to wild-type vacuoles (Figs. 1 and 2; ref. 31) or through genetic overexpression of all four vacuolar SNAREs (Figs. 3–5). Lysis and fusion driven by excess SNAREs can be independent of Ypt7p (Fig. 7).
Both lysis and membrane fusion entail localized lipid restructuring to nonbilayer configurations. Our current assay system, which measures luminal content mixing and release, shows that both fusion and lysis require SNARE function and occur as a final kinetic step, well after trans-SNARE complex formation (Fig. 2C). Our fusion and lysis values may be underestimates as our assays will only measure the initial round of vacuole lysis, would not measure the fusion of a vacuole bearing proPho8p with a vacuole that had previously lysed and thereby spilled its luminal proteases, and do not measure fusion events between pairs of vacuoles lacking proteases or pairs of vacuoles lacking proPho8p, as noted (29). Nonetheless, the rates of both fusion and lysis of 4SNARE++ vacuoles are as enhanced by Sec18p at the earliest incubation times, when little lysis or fusion has occurred, as at later times (Fig. 5 C and D).
Liposomes bearing biologically relevant v-SNARE and t-SNARE complexes in separate vesicles exhibit SNARE-mediated lipid mixing (12) and substantial SNARE-induced lysis followed by membrane reannealing (14, 15), although fusion does occur in those liposomes that remain sealed (13, 39). Our studies show that Rab-dependent SNARE pairing can induce either lysis or fusion of an organelle membrane. Physiological levels of Ypt7p, HOPS, and vacuolar SNAREs support efficient fusion with minimal Rab- and SNARE-dependent lysis, although a background of SNARE-independent lysis is seen in vitro. Vam7p, which stimulates trans-SNARE pairing (31), stimulates the lysis of normal vacuoles (Fig. 1B). When all of the SNAREs are overproduced, lysis only requires added Sec18p (Figs. 4A and 5D). Regulatory lipids are essential for concentrating the wild-type levels of SNAREs at the vertex ring (3), but when the SNAREs are overexpressed and liberated from cis-SNARE complexes by Sec18p (Fig. 4B), they may achieve sufficient vertex ring levels for fusion without Ypt7p-dependent and regulatory lipid-dependent concentration.
Is Rab regulation negative, in the sense of an on/off switch that either blocks or permits an otherwise robust fusion activity that is inherent to the SNAREs, or positive, through activating an otherwise very slow fusion to occur at rates that are biologically meaningful? The fusion of wild-type vacuoles is supported by the HOPS/SNARE complex (24), and each step of docking is reversible until fusion occurs (40). Although only a few percent of the vacuole SNAREs enter trans complexes, increasing the vacuolar level of either Nyv1p, Vam7p, or HOPS enhances trans-SNARE complex assembly (31). These findings suggest that the rate of formation of HOPS/trans-SNARE complex is limiting for fusion, rather than complex formation being limited by the stoichiometry of supply of one of its five constituents. Rab family GTPases are thought to regulate the formation of SNARE complexes (24, 41), the committed step of the pathway to fusion. An apparent rate constant k can be defined for the fifth-order association of HOPS, Vam3p, Vam7p, Vti1p, and Nyv1p. We find comparable rates of fusion in two conditions: unsupplemented reactions with vacuoles bearing wild-type levels of Ypt7p, HOPS, and SNAREs and reactions with 4SNARE++ ypt7Δ vacuoles supplemented with Sec18p and an excess of HOPS. This finding suggests that the rates of forming the fusion-competent HOPS/SNARE complex are equal under each condition. Using values of SNARE overexpression from Fig. 3B,
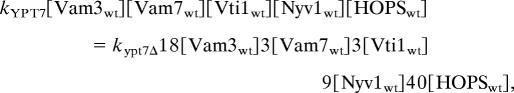 |
and kYPT7/kypt7Δ = 58,320, i.e., between 104 and 105. This calculation rests on several assumptions, such as the overexpressed SNAREs being fully active and accessible for trans-complex assembly, and we note that 4SNARE++ ypt7Δ vacuole fusion with Sec18p added alone is approximately half that seen with added Sec18p and HOPS. Nevertheless, our experiments and this simple calculation illustrates that modest increases in each of five factors can drive their assembly into a complex without Rab catalysis. Although the means whereby Ypt7p so dramatically promotes HOPS/SNARE complex assembly are unknown, Ypt7p is required for the assembly of the SNAREs, HOPS, and regulatory lipids into a functional, localized vacuole microdomain, the vertex ring (3, 5, 6). The delivery of other proteins and lipids to the vacuole or their spatial enrichment at the vertex ring microdomain may also be regulated by Ypt7p, which may contribute to the substantial effect of Ypt7p on the apparent rate constant of HOPS/SNARE complex formation. We suggest that this spatial concentration of the SNAREs and HOPS is a major factor in the catalysis of functional HOPS/SNARE complex assembly by Ypt7p. Just as a part of enzymic catalysis can occur through binding inherently reactive substrates at adjacent sites on the enzyme surface (42), so the Rabs concentrate multiple SNAREs and key lipids next to each other (in the vertex ring) and across from each other (by aligning the tethered rings of each vacuole), which may be a major part of how Ypt7p catalyzes the assembly of HOPS/trans-SNARE complex. Our data are also consistent with Ypt7p activating the fusion capacity of assembled HOPS/SNARE complexes or even participating more directly in the fusion reaction.
Lysis may be mediated by excess trans-SNARE complexes at the vertex ring. When Ypt7p-directed vertex ring assembly is intact, overexpression of SNAREs gives substantial lysis (Figs. 4 and 5). When these vacuoles undergo extraction of Ypt7p by Gyp1–46 and Gdi1p, lysis is reduced to baseline levels, whereas there is only a 40% reduction of fusion (Fig. 4D), consistent with a higher level of SNARE pairs being needed for lysis than for fusion. The same levels of SNARE enrichment at the vertex ring, trans-SNARE assembly, and fusion may be achieved by either Ypt7p-driven enrichment at wild-type SNARE levels or SNARE overexpression in the absence of a Ypt7p-driven enrichment mechanism. In accord with the concept of SNARE complex assembly without Ypt7p, the cytosolic domains of the three Q-SNAREs can associate with Nyv1p without the aid of Ypt7p (40). Furthermore, ypt7Δ vacuoles cannot fuse at all (data not shown), whereas 4SNARE++ ypt7Δ vacuoles show moderate fusion when given Sec18p alone (Fig. 7) and require the further addition of Vam7p, HOPS, or both for optimal fusion. These 4SNARE++ ypt7Δ vacuoles give lower levels of lysis than are seen with vacuoles from isogenic YPT7 strains.
What is the order of late events in vacuole fusion? Reactions that are synchronized (28, 29) by adding Vam7p rapidly become resistant to antibody to Vam3p, indicating trans-SNARE pairing, followed almost at once by lipid mixing (18) and Ca2+ efflux from the vacuole lumen (28). We suggest that lipid mixing and a partial efflux of luminal Ca2+ stores reflect hemifusion around the vertex ring. Lipid rearrangements or other reactions required for completion of fusion or lysis are far slower; further study is needed to determine all of the factors that regulate the crucial choice between fusion and lysis. HOPS shares with synaptotagmin the property of physically associating with 4SNARE complexes for fusion, and both have lipid-interacting properties; they may each favor fusion over lysis.
In addition to providing localized stress on the bilayer, trans-SNARE complexes may contribute to fusion by other means. Syntaxin and synaptobrevin transmembrane domains destabilize bilayers (10). The clustering of these domains, which is inherent in the assembly of trans-SNARE complexes, may localize their bilayer-destabilization function. Vacuole fusion requires several regulatory lipids (diacylglycerol, phosphoinositides, and ergosterol), which require Ypt7p, HOPS, and SNAREs for their mutual vertex enrichment. Lipids such as diacylglycerol are themselves highly fusogenic (43); SNAREs and HOPS may contribute to fusion by promoting their selective localization. Finally, there is a dramatic break in the curvature of the membranes at the vertex ring, where the flat boundary membrane meets the gently curved outside membrane, and bilayer structure may be destabilized by this curvature change.
Materials and Methods
Strains, Plasmids, and Genetic Manipulations.
Yeast strains BJ3505 [MATα ura3-52 trp1-Δ101 his3-Δ200 lys2-801 gal2 (gal3) can1 prb1-Δ1.6R pep4::HIS3] (44) and DKY6281 (MATα ura3-52 leu2-3,112 trp1-Δ901 his3-Δ200 lys2-801 suc2-Δ9 pho8::TRP1) (17) were used for vacuole production and genetic constructions.
The plasmid pYJ406 was generated by introducing the 720-bp upstream region and the 300-bp downstream region of ADH1 into the XhoI/HindIII and SacI/SacII sites of pRS406. Likewise, pYJ403, pYJ404, and pYJ405 were generated from pRS403, pRS404, and pRS405. The plasmid pYJ406-VAM3 for overexpression of Vam3p under the ADH1 promoter was generated by introducing the ORF and the 311-bp downstream region of VAM3 into the BamHI/SacII sites of pRS406. The resulting pYJ406-VAM3 lacks the 300-bp downstream region of ADH1. pYJ404-VTI1 and pYJ405-VTI1 were constructed by ligating the ORF and the 208-bp downstream region of VTI1 into BamHI/SacII sites of pYJ404 and pYJ405. pYJ403-VAM7 was constructed by introducing the ORF and the 150-bp downstream region of VAM7 into the BamHI/SacII sites of pYJ403. pYJ400-VAM7 was generated by ligating the XhoI/SacII-digested fragment that contains the ADH1 promoter region followed by the VAM7 gene from pYJ403-VAM7 with the XhoI/SacII-digested pRS400. pYJ406-NYV1 was generated by subcloning the NYV1 ORF and 633-bp downstream region into the BamHI/SacII sites of pYJ406. pYJ408-NYV1 was constructed by introducing the XhoI/SacII fragment of pYJ406-NYV1 containing the ADH1 promoter region followed by NYV1 into the XhoI/SacII sites of pRS408 (a generous gift from Frederick Cross, The Rockefeller University, New York, NY). To target GFP to the vacuole, a DNA fragment encoding the signal peptide of the MEL1 gene, which encodes α-galactosidase in Saccharomyces carlsbergensis (45), was fused in-frame to the 5′ end of the gene encoding Superglow GFP (sGFP) in pYJ406 or pYJ400, generating pYJ406-mel-sGFP or pYJ400-mel-sGFP.
To generate yeast strains overexpressing vacuolar SNAREs under control of the ADH1 promoter, BJ3505 and DKY6281 were sequentially transformed with BsaBI-digested pYJ406-VAM3, NdeI-linearized pYJ404-VTI1 (for BJ3505), or pYJ405-VTI1 (for DKY6281), and BsaBI-digested pYJ400-VAM7. The resulting BJ3505 three Q-SNARE++ and DKY6281 three Q-SNARE++ strains were transformed with BsaBI-linearized pYJ408-NYV1 to generate BJ3505 and DKY6281 overexpressing four vacuolar SNARE proteins (BJ3505 4SNARE++ and DKY6281 4SNARE++), respectively. To generate yeast strains that overexpress Nyv1p (BJ3505 NYV1++ and DKY6281 NYV1++), BJ3505 and DKY6281 were transformed with BsaBI-linearized pYJ406-NYV1. The YPT7 gene was deleted in BJ3505 4SNARE++ and DKY6281 4SNARE++ by using PCR-based gene disruption (46), generating BJ3505 4SNARE++ ypt7Δ and DKY6281 4SNARE++ ypt7Δ, respectively.
To generate yeast strains containing vacuoles with luminal GFP, BJ3505, BJ3505 4SNARE++, and BJ3505 4SNARE++/ypt7Δ strains were transformed with BsaBI-linearized pYJ406-mel-sGFP. sGFP expression and vacuolar localization were confirmed by immunoblotting and fluorescence microscopy.
Vacuole Isolation and in Vitro Vacuole Fusion Assay.
Vacuoles were isolated as described (17). Yeast strains overproducing the four SNAREs were grown on complete synthetic medium lacking histidine, leucine, lysine, tryptophan, and uracil, supplemented with 0.1% sodium glutamate as nitrogen source, G418 sulfate (200 μg/ml), and CloNAT (100 μg/ml). For strains in DKY6281 background, histidine (0.002%) and lysine (0.003%) were added. To purify vacuoles, yeast were subcultured from a log-stage minimal culture into rich medium (yeast extract/peptone/dextrose) for overnight growth (≈9–11 generations) without selection; genetic marker loss was <3%.
Standard 30-μl in vitro fusion reactions at 27°C contained 20 mM Pipes-KOH (pH 6.8), 200 mM sorbitol, 125 mM KCl, 6 mM MgCl2, 1 mM ATP, 1 mg/ml creatine kinase, 29 mM creatine phosphate, 10 μM CoA, 815 nM purified Pbi2p (I2B), 3 μg pep4Δ vacuoles (from BJ3505 derivatives), and 3 μg pho8Δ vacuoles (from DKY6281 derivatives). Pho8p phosphatase activity was assayed as a measure of vacuole fusion (47). Fusion units are μmol p-nitrophenylate formed min−1·μg−1 pep4Δ vacuole.
Reagents.
Antibodies were prepared as described and dialyzed into PS buffer (10 mM Pipes/KOH, pH 6.8/200 mM sorbitol) with 125 mM KCl. Concentrations used (unless otherwise noted) were: 209 nM affinity-purified anti-Sec17p antibody (48), 467 nM affinity-purified anti-Ypt7p antibody (49), and 900 nM anti-Vam3p (6). The antigenic peptide for α-Ypt7p antibody (49) was dissolved in PS buffer and added to reactions at a final concentration of 67 μg/ml.
Purified recombinant proteins were dialyzed into PS buffer with 125 mM KCl and used at the following concentrations: 0.6 μM his6-Gyp1–46p (6), 1.2 μM GST-FYVE domain (50), 10 μM C1b domain (51), 36 μM GST-ENTH domain (52), 10 μM MARCKS effector domain (53), 95 nM gel-filtered his6-Sec18p (33), and 10.5 μg/ml HOPS (20).
A plasmid-expressing recombinant Vam7p-CBD fusion protein was a generous gift from Alexey Merz (University of Washington, Seattle, WA). Recombinant Vam7p, purified by chitin affinity chromatography and intein cleavage, was stored in 20 mM Hepes-NaOH, pH 8.0, and 300 mM NaCl and routinely added to fusion reactions at 50 nM.
GDI1 was PCR-amplified from yeast by using the primers GGTGGTCATATGGATCAAGAAACAATAG and GGTGGTCCCGGGCTGCTTTTCTTGTTCTTG. The PCR fragment was digested with NdeI and SmaI and ligated into pTYB3 (New England Biolabs, Ipswich, MA) digested with the same enzymes, creating pVJS3. This plasmid, transformed into Escherichia coli Rosetta 2 (λDE3) (Novagen, San Diego, CA), expressed Gdi1p-CBD fusion protein that was purified by chitin affinity chromatography and intein cleavage. Purification followed the manufacturer's directions, except that Triton X-100 was omitted from the wash and elution steps. Purified rGdi1p was dialyzed into PS buffer with 125 mM KCl. rGdi1p contained an additional C-terminal glycyl residue that did not affect its activity (data not shown). Gdi1p was routinely added to fusion reactions at 1.2 μM in the absence of Gyp1–46p and 0.6 μM when used with Gyp1–46p.
GFP Release (Lysis) Assay.
Vacuoles containing luminal sGFP were isolated from the protease-deficient BJ3505 background and premixed in a 1:1 (fluorescent pep4Δ/nonfluorescent pho8Δ) ratio before the addition of other reaction components. Standard vacuole fusion reactions were used for the GFP release assay, with the following modifications: reactions contained 0.1× protease inhibitor mixture (50× stock: 13 μg/ml leupeptin, 25 mM 1,10-phenanthroline, 25 μg/ml pepstatin A, and 5 mM Pefabloc SC) to stabilize GFP after vacuolar release. Each reaction was performed on a 3× scale (90 μl).
At specified times, 30-μl aliquots were moved to a prechilled tube and kept at 0°C until assayed for Pho8p activity (fusion). For sGFP detection, 30-μl aliquots were placed in a prechilled tube containing 30 μl of PS buffer and reaction salt (20 mM Pipes-KOH, pH 6.8/200 mM sorbitol/125 mM KCl/5 mM MgCl2). Samples were centrifuged (5,200 × g, 4°C, 6 min; no further sedimentation of GFP was seen at higher centrifugal speeds), and 50 μl of the supernatants was mixed with 10 μl of chilled supernatant dilution buffer (200 mM sorbitol/20 mM Pipes-KOH pH 6.8, 125 mM KCl, 5 mM MgCl2, 3% Triton X-100). Vacuolar membrane pellets were resuspended in 59 μl of solubilization buffer (200 mM sorbitol/20 mM Pipes-KOH, pH 6.8/125 mM KCl/5 mM MgCl2/0.5% Triton X-100/1× protease inhibitor mixture). Twenty microliters of each pellet/supernatant sample was placed into a 384-well black, low-volume/round bottom plate (Corning, Corning, NY). Fluorescence was read in a SpectraMAX Gemini XPS plate reader (Molecular Devices, Sunnyvale, CA) (λex = 462 nm, λem = 510 nm, emission cutoff = 495 nm), with 30 reads per well on the “high” PMT setting. Total GFP was determined from vacuoles solubilized in a final volume of 60 μl. Raw supernatant fluorescence readings were multiplied by 1.2 to compensate for dilution, and these values were added to the pellet value to determine total GFP recovered from each reaction.
Acknowledgments
We thank Christopher Hickey (Dartmouth Medical School) for providing purified HOPS, Rutilio Fratti (University of Illinois at Urbana-Champaign, Urbana, IL) for SD and PX domain proteins, and Nathan Margolis and Naomi Thorngren for expert assistance. This work was supported by National Institutes of Health Grant GM23377. V.J.S. was supported by Damon Runyon Foundation Fellowship DRG-1837 and National Institutes of Health Autoimmunity and Connective Tissue Training Grant T32 AR07576 (to Dartmouth Medical School).
Abbreviations
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- HOPS
homotypic fusion and vacuole protein sorting complex
- sGFP
Superglow GFP.
Note Added in Proof.
Lysis from an imbalance of membrane fusion catalysts is also seen in cell:cell fusion (54).
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13541.
References
- 1.Jahn R, Lang T, Südhof TC. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 2.Grosshans BL, Ortiz D, Novick P. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. J Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miaczynska M, Zerial M. Exp Cell Res. 2002;272:8–14. doi: 10.1006/excr.2001.5401. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Seeley ES, Wickner W, Merz AJ. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Merz AJ, Collins KM, Wickner W. J Cell Biol. 2003;160:365–374. doi: 10.1083/jcb.200209095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess SW, McIntosh TJ, Lentz BR. Biochemistry. 1992;31:2653–2661. doi: 10.1021/bi00125a004. [DOI] [PubMed] [Google Scholar]
- 8.Haque ME, McIntosh TJ, Lentz BR. Biochemistry. 2001;40:4340–4348. doi: 10.1021/bi002030k. [DOI] [PubMed] [Google Scholar]
- 9.Skehel JJ, Wiley DC. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 10.Langosch D, Crane JM, Brosig B, Hellwig A, Tamm LK, Reed J. J Mol Biol. 2001;311:709–721. doi: 10.1006/jmbi.2001.4889. [DOI] [PubMed] [Google Scholar]
- 11.Jin H, Carlile C, Nolan S, Grote E. Eukaryot Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 13.Nickel W, Weber T, McNew JA, Parlati F, Söllner TH, Rothman JE. Proc Natl Acad Sci USA. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennison SM, Bowen ME, Brunger AT, Lentz BR. Biophys J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Arac D, Wang T-M, Gilpin CJ, Zimmerberg J, Rizo J. Biophys J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Nat Struct Mol Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 17.Haas A, Conradt B, Wickner W. J Cell Biol. 1994;126:87–97. doi: 10.1083/jcb.126.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jun Y, Wickner W. Proc Natl Acad Sci USA. 2007 in press. [Google Scholar]
- 19.Mayer A, Wickner W. J Cell Biol. 1997;136:307–317. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stroupe C, Collins KM, Fratti RA, Wickner W. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaGrassa TJ, Ungermann C. J Cell Biol. 2005;168:401–414. doi: 10.1083/jcb.200407141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seals D, Eitzen G, Margolis N, Wickner W, Price A. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dulubova I, Yamaguchi T, Wang Y, Südhof TC, Rizo J. Nat Struct Biol. 2001;8:258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
- 24.Collins KM, Thorngren NL, Fratti RA, Wickner WT. EMBO J. 2005;24:1775–1786. doi: 10.1038/sj.emboj.7600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Nat Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 26.Maier K, Muller H, Holzer H. J Biol Chem. 1979;254:8491–8497. [PubMed] [Google Scholar]
- 27.Peters C, Mayer A. Nature. 1998;396:575–580. doi: 10.1038/25133. [DOI] [PubMed] [Google Scholar]
- 28.Merz AJ, Wickner WT. J Cell Biol. 2004;164:195–206. doi: 10.1083/jcb.200310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merz AJ, Wickner WT. Proc Natl Acad Sci USA. 2004;101:11548–11553. doi: 10.1073/pnas.0404583101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eitzen G, Will E, Gallwitz D, Haas A, Wickner W. EMBO J. 2000;19:6713–6720. doi: 10.1093/emboj/19.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins KM, Wickner WT. Proc Natl Acad Sci USA. 2007;104:8755–8760. doi: 10.1073/pnas.0702290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fratti RA, Collins KM, Hickey CM, Wickner W. J Biol Chem. 2007;282:14861–14867. doi: 10.1074/jbc.M700971200. [DOI] [PubMed] [Google Scholar]
- 33.Thorngren N, Collins K, Fratti R, Wickner W, Merz AJ. EMBO J. 2004;23:2765–2776. doi: 10.1038/sj.emboj.7600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ungermann C, Price A, Wickner W. Proc Natl Acad Sci USA. 2000;97:8889–8891. doi: 10.1073/pnas.160269997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dascher C, Ossig R, Gallwitz D, Schmitt HD. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lian JP, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Nature. 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- 37.Kendall DA, MacDonald RC. J Biol Chem. 1982;257:13892–13895. [PubMed] [Google Scholar]
- 38.Lau WL, Ege DS, Lear JD, Hammer DA, DeGrado WF. Biophys J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Zhang F, Su Z, McNew JA, Shin Y-K. Nat Struct Mol Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 40.Jun Y, Thorngren N, Starai VJ, Fratti RA, Collins K, Wickner W. EMBO J. 2006;25:5260–5269. doi: 10.1038/sj.emboj.7601413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Søgaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirschhausen T, Rothman JE, Söllner T. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 42.Narlikar GJ, Herschlag D. Annu Rev Biochem. 1997;66:19–59. doi: 10.1146/annurev.biochem.66.1.19. [DOI] [PubMed] [Google Scholar]
- 43.Allan D, Thomas P, Mitchell RH. Nature. 1978;276:289–290. doi: 10.1038/276289a0. [DOI] [PubMed] [Google Scholar]
- 44.Jones E. Methods Enzymol. 2002;351:127–150. doi: 10.1016/s0076-6879(02)51844-9. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Xu H, Bentley WE, Rao G. Biotechnol Prog. 2002;18:831–838. doi: 10.1021/bp020066t. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein AL, McCusker JH. Yeast. 1999;15:1541–1555. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 47.Haas A. Methods Cell Sci. 1995;17:283–294. [Google Scholar]
- 48.Haas A, Wickner W. EMBO J. 1996;15:3296–3305. [PMC free article] [PubMed] [Google Scholar]
- 49.Eitzen G, Thorngren N, Wickner W. EMBO J. 2001;20:5650–5656. doi: 10.1093/emboj/20.20.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. EMBO J. 2000;19:5012–5019. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson JE, Giorgione J, Newton AC. Biochemistry. 2000;39:11360–11369. doi: 10.1021/bi000902c. [DOI] [PubMed] [Google Scholar]
- 52.Hyman J, Chen H, DiFiore PP, De Camilli P, Brunger AT. J Cell Biol. 2000;149:537–546. doi: 10.1083/jcb.149.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Arbuzova A, Hangyas-Mihalyne G, McLaughlin S. J Biol Chem. 2001;276:5012–5019. doi: 10.1074/jbc.M008355200. [DOI] [PubMed] [Google Scholar]
- 54.Aguilar PS, Engel A, Walter P. Mol Biol Cell. 2007;18:547–556. doi: 10.1091/mbc.E06-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]