Abstract
HIV-1 integrase, the viral enzyme responsible for provirus integration into the host genome, can be actively degraded by the ubiquitin–proteasome pathway. Here, we identify von Hippel–Lindau binding protein 1(VBP1), a subunit of the prefoldin chaperone, as an integrase cellular binding protein that bridges interaction between integrase and the cullin2 (Cul2)-based von Hippel–Lindau (VHL) ubiquitin ligase. We demonstrate that VBP1 and Cul2/VHL are required for proper HIV-1 expression at a step between integrase-dependent proviral integration into the host genome and transcription of viral genes. Using both an siRNA approach and Cul2/VHL mutant cells, we show that VBP1 and the Cul2/VHL ligase cooperate in the efficient polyubiquitylation of integrase and its subsequent proteasome-mediated degradation. Results presented here support a role for integrase degradation by the prefoldin–VHL–proteasome pathway in the integration–transcription transition of the viral replication cycle.
Keywords: prefoldin, ubiquitin, VHL, retrovirus, transcription
Integration of the HIV-1 genome into host chromosomes constitutes a crucial step for productive infection by retroviruses (1). After cell entry and uncoating of the viral capsid, the genomic HIV-1 RNA is reverse-transcribed into linear dsDNA that assembles with cellular and viral proteins including integrase (IN) to form the preintegration complex. Nuclear import of this large nucleoprotein complex allows the viral DNA to integrate into host chromosomes, thus leading to transcription and expression of viral genes. Although IN has been reported to participate in nonintegrative steps of the viral replication cycle, such as reverse transcription (2–6), viral DNA nuclear import (7–9), and viral particle production (10), IN undoubtedly catalyzes viral genome integration. IN forms a tetramer stably associated with a pair of viral DNA ends (11) and catalyzes two distinct steps of the integration process. The first step, called 3′ processing, corresponds to the removal of two nucleotides from each 3′ end of the viral DNA (1, 12) and precedes the strand-transfer reaction in which the 3′-processed viral DNA ends are covalently joined to the target DNA (13). The integration process is finally accomplished by cleavage of unpaired dinucleotides from the 5′ ends of viral DNA and repair of single-stranded gaps created by the strand-transfer reaction between viral and target DNA (14–16). Unlike 3′ processing and strand-transfer reactions, gap repair is not mediated by IN but has rather been proposed to be carried out by host-DNA repair enzymes that are not yet clearly defined. Furthermore, it has been proposed that gap repair requires active disassembly of IN from strand-transfer products by as of yet unknown mechanisms (11, 17).
To identify cellular factors that participate in or interfere with viral integration, cellular proteins interacting with IN have been screened for and characterized (18–22). In particular, the transcriptional coactivator lens epithelium-derived growth factor/transcription coactivator p75 (LEDGF/p75) has been reported to tether IN to chromosomes (23, 24) and to contribute to the targeting of viral DNA to preferential integration sites (25). Other cellular proteins also participate in the HIV-1 integration process in the context of a host-cell infection but still await further characterization (26, 27).
IN has been shown to be actively degraded by the ubiquitin–proteasome pathway (24, 28–30). Ubiquitin conjugation is accomplished through an enzymatic cascade with ubiquitin first being activated by a unique E1 enzyme, transferred from E1 to an E2 ubiquitin conjugating enzyme, which then transfers the ubiquitin to a lysine residue on the substrate in conjunction with an E3 ubiquitin-protein ligase that provides substrate specificity. Two main classes of E3 ligases have been characterized: homologous to E6-AP C terminus (HECT)-type E3s display catalytic activity, whereas single or multisubunit RING-H2-type E3s promote ubiquitinylation by positioning the activated E2 in close proximity to the substrate. Cullin-RING complexes comprise the largest known class of ubiquitin ligases. Ubiquitin may be attached to proteins as a monomer or as polymers that lead to many distinct functions, but lysine 48-linked polyubiquitin chains promote recognition by the 26S proteasome and degradation of the polyubiquitylated protein. HIV-1-processed IN, excised from the Gag-Pol polyprotein by the viral protease, presents an N-terminal phenylalanine, which serves as a degradation signal also called N-degron, recognized by the N-end rule ubiquitin–proteasome degradation pathway (30). When this phenylalanine is preceded by a methionine, thereby masking the N-end rule degradation signal, or in cells depleted for the N-end rule-specific ubiquitin ligases UBR1, 2 and 4, an alternative but yet uncharacterized pathway also leads to IN ubiquitin-mediated degradation (24, 28, 29, 31). However, the precise function of this active IN turnover in the viral life cycle has not been elucidated so far.
In this article, we identify an IN-interacting protein, von Hippel–Lindau binding protein 1 (VBP1), a component of the prefoldin chaperone, and we provide evidence indicating that VBP1 targets IN for cullin2 (Cul2)-von Hippel–Lindau (VHL) ubiquitin ligase-mediated polyubiquitylation and subsequent degradation by the 26S proteasome. We also show that VBP1 is involved in HIV-1 gene expression after the strand-transfer reaction where it is required to allow proper transcription of the viral genome.
Results
VBP1 Is an Integrase Cellular-Binding Partner.
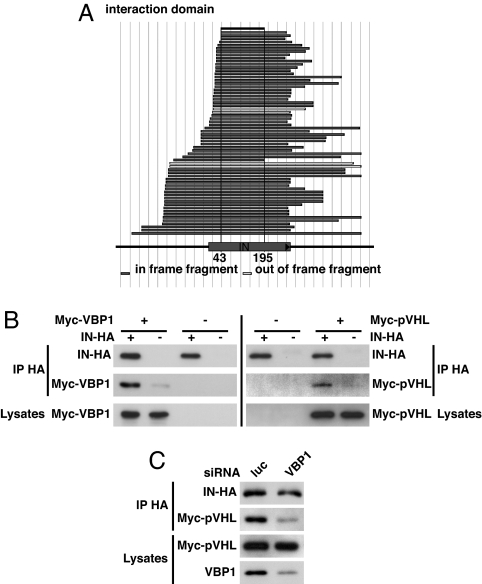
To identify cellular proteins that participate in or interfere with IN-mediated HIV-1 integration into the host genome, we used HIV-1 YU2 IN as a bait in a yeast two-hybrid assay to screen two highly complex cDNA libraries generated by using the human T lymphoblastic (CEM T) cell line. By this approach, multiple overlapping fragments of the VBP1 gene were identified [10 different fragments with an oligo(dT)-primed library and 1 fragment with a random-primed library], mostly containing the full-length VBP1 coding sequence (data not shown). Conversely, screening a highly complex library of HIV-1 random fragments by using VBP1 as bait led exclusively to the identification of the IN gene, indicating that IN is the unique target of VBP1 within the HIV-1 viral genome. In addition, alignment of the IN-interacting fragments revealed that IN binds VBP1 through residues 43–195 of IN, a region mainly included in its catalytic core domain (Fig. 1A).
Fig. 1.
VBP1 specifically interacts with HIV-1 IN and mediates IN–pVHL interaction. (A) A yeast two-hybrid screening of a highly complex library of HIV-1 random fragment was performed by using VBP1 as bait. All obtained clones contained the IN gene, and their alignment allows the mapping of a VBP1 interacting domain in IN sequence located between residues 43 and 195. (B) HeLa cells or IN-HA cells (HeLa cells stably expressing IN-HA) were transfected with Myc-VBP1 or Myc-pVHL expression plasmids. Equal amounts of total cellular proteins (lysates) were immunoprecipitated by using anti-HA antibody. Immunoprecipitated proteins were then analyzed by Western blotting with anti-HA or anti-Myc antibody. (C) IN-HA cells were transfected with VBP1-specific (VBP1) or control luciferase (luc)-directed siRNA, prior to transfection with Myc-pVHL expression plasmid. After immunoprecipitation with anti-HA antibody, immunoprecipitates, as well as endogenous VBP1 and transfected Myc-pVHL in cell lysates, were analyzed by Western blotting by using anti-HA, anti-Myc, or anti-VBP1 antibodies, as indicated. Signals were quantified by densitometric analysis of the scanned autoradiographic films by using ImageJ software and revealed a 69% decrease of the coimmunoprecipitated Myc-pVHL/immunoprecipitated IN-HA ratio in the VBP1 siRNA-treated cells compared with the control cells.
Specific interaction between IN and VBP1 was further analyzed by coimmunoprecipitation assays in cells expressing both HA-tagged IN (IN-HA) and Myc-tagged VBP1 (Fig. 1B Left). For this purpose, HeLa cells stably expressing an HA-tagged version of IN were transiently transfected with an Myc-VBP1 expression vector. In agreement with the two-hybrid data, Myc-VBP1 specifically coimmunoprecipitated with IN-HA (Fig. 1B Left), thus clearly indicating the interaction of HIV-1 IN with a previously unrecognized cellular partner, VBP1.
VHL Protein (pVHL) Interacts with Integrase in a VBP1-Dependent Manner.
VBP1 was initially characterized as a partner of pVHL, the substrate recognition component of the Cul2/VHL ubiquitin ligase complex, also composed of elongin C, elongin B, Cul2, and the RING finger protein, Rbx1 (32, 33) (data not shown).
Coimmunoprecipitation assays showed that transiently expressed Myc-pVHL specifically interacted with IN-HA (Fig. 1B Right). This interaction was significantly reduced upon siRNA-based inactivation of VBP1 endogenous expression, thus indicating that IN-pVHL interaction requires VBP1 (Fig. 1C).
These results demonstrate that IN interacts with both VBP1 and the ubiquitin ligase complex component pVHL, and that interaction with pVHL is bridged by VBP1.
VBP1-Containing Prefoldin and VHL Ubiquitin Ligase Participate in HIV-1 Gene Expression.
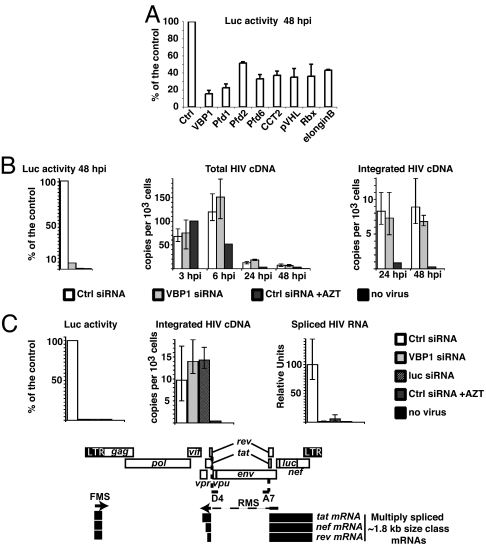
To address the potential role of VBP1 and pVHL during HIV-1 replication, we analyzed the effect of VBP1 or pVHL knockdown on HIV-1 replication in a single-round assay where HeLa cells were infected with a NL4-3Δenv virus pseudotyped with VSV-G envelope and containing the luciferase gene inserted into the nef gene (NL4-3ΔenvLuc VSVg). HeLa cells were first transfected with siRNA and infected, and luciferase activity was measured 48 h after infection. In this assay, reduction of VBP1 or pVHL expression by specific siRNAs resulted in a highly significant decrease in luciferase activity. Compared with cells treated with a control nontargeting siRNA, luciferase activity decreased 3.7- to 12-fold by using two different siRNA directed against VBP1 and 2.8-fold with pVHL-directed siRNA (Fig. 2A and data not shown). These results indicate that VBP1 and pVHL are involved in HIV-1 replication at a step(s) occurring after viral entry up to and including translation of Nef-coding mRNA.
Fig. 2.
VBP1 and the Cul2/VHL ligase are important for HIV-1 gene expression at a postintegration step. (A) HeLa cells were transfected with siRNAs directed against indicated targets or control nontargeting (Ctrl) siRNA. Cells were subsequently infected with NL4-3ΔenvLuc VSVg virus, and luciferase activity was monitored in cell lysates 48 h after infection. (B) HeLa cells were transfected with either VBP1-directed (VBP1) or control nontargeting siRNA as indicated. Cells were subsequently infected with NL4-3ΔenvLuc VSVg virus or mock-infected, in the presence or absence of 3′-azido-3′-deoxythymidine, and luciferase activity was monitored in cell lysates. At different time points, DNA was extracted and subjected to real-time PCR analysis to quantify total viral DNA and integrated proviral DNA. (C) HeLa cells were transfected with either VBP1-directed, luciferase (luc)-directed, or control nontargeting siRNA as indicated. Cells were subsequently infected with NL4-3ΔenvLuc VSVg virus or mock-infected, in the presence or absence of 3′-azido-3′-deoxythymidine, and luciferase activity was monitored in cell lysates. Both DNA and RNA were extracted from each sample and subjected to real-time PCR or RT-PCR, respectively. Multiply spliced viral mRNAs (≈1.8 kb size class viral mRNAs) were selectively amplified by using a reverse primer encompassing the junction between the donor D4 and acceptor A7 splice sites involved in the formation of tat, rev, and nef mRNA. Integrated provirus and multiply spliced mRNAs were quantified for each sample. The scheme under the graphs represents the genetic organization of the HIV-1 genome and the ≈1.8-kb size class viral transcripts tat, rev, and nef (black bars) selectively amplified with the forward multiply spliced (FMS) and reverse multiply spliced (RMS) primers (arrows).
In addition to its ability to bind pVHL, VBP1 also functions as a subunit of the heterohexameric molecular chaperone prefoldin with VBP1 being identified as prefoldin 3 (34). The prefoldin complex binds to nonnative target proteins, such as actin and tubulin proteins, and transfers them to another chaperone, the cytosolic chaperonin CCT2 (chaperonin containing TCP-1, subunit 2; also termed c-cpn or TriC), which facilitates their correct folding (34, 35). To investigate the influence of prefoldin and CCT2 chaperones on HIV-1 replication, expression of different subunits of prefoldin or CCT2 was knocked down by using specific siRNAs before infection with NL4-3ΔenvLuc VSVg virus. As shown in Fig. 2A, affecting the expression of prefoldin and CCT2 decreased HIV-1 gene expression by 50–85% as measured by luciferase activity.
To determine whether pVHL was involved in HIV-1 replication as a component of the Cul2/VHL ligase complex, we analyzed the effect of a decreased expression of the Cul2/VHL ligase complex on HIV-1 gene expression and found that silencing Rbx1 and elongin B expression also reduced luciferase activity by 55–65% of the control (Fig. 2A).
These results thus indicate that interaction between IN, VBP1, and pVHL as identified by two-hybrid and coimmunoprecipitation approaches is relevant in the context of HIV-1 replication and that both prefoldin and Cul2/VHL ubiquitin ligase complexes are required for proper HIV-1 gene expression.
VBP1 Controls HIV-1 Expression at a Postintegration Step.
To further characterize the step of the virus life cycle affected by VBP1 knockdown, we measured the levels of total HIV-1 DNA and integrated forms of proviral DNA by quantitative PCR using cell extracts from siRNA-transfected cells subsequently infected with NL4-3ΔenvLuc VSVg virions. As shown in Fig. 2B, VBP1-targeting siRNA did not significantly affect the amount of total reverse-transcribed HIV-1 cDNA nor integrated provirus, compared with control siRNA, whereas a 14-fold reduction of luciferase activity was measured after VBP1 knockdown. These data, therefore, suggest that VBP1 is involved after the strand-transfer step of proviral DNA integration.
We next evaluated the impact of VBP1 knockdown on the transcriptional activity of integrated provirus. Cells were first transfected with siRNA and subsequently infected with NL4-3ΔenvLuc VSVg virions. Forty-eight hours after infection, integrated proviral DNA was quantified for each sample by real-time PCR and the expression level of multiply spliced (≈1.8 kb size class) viral mRNAs was quantified by real-time RT-PCR (Fig. 2C). As expected, inhibition of reverse transcriptase by 3′-azido-3′-deoxythymidine resulted in the inhibition of the integration of both proviral DNA and its subsequent transcription. Compared with a control siRNA, an siRNA targeting the luciferase gene carried by the virus did not affect proviral integration but led to a strong decrease of multiply spliced viral mRNAs because of the siRNA-mediated degradation of newly synthesized viral transcripts. Importantly, VBP1 knockdown almost completely inhibited expression of viral multiply spliced RNA with a decrease of 72-fold, although no difference in the amount of integrated provirus was detected (Fig. 2C). VBP1 is, therefore, required for the proper transcription of viral genes by acting after the strand-transfer step of proviral DNA integration.
Integration Is Required Before Transcriptional Control of HIV-1 by VBP1.
To analyze whether VBP1 controls the transition between integration of the viral DNA into the host genome and transcription or rather exerts a general effect on the transcription process, we first compared the effect of siRNAs on viral genome expression either after single-round infection and genome integration or after genome transfection. For this purpose, siRNA-transfected HeLa cells were either infected with HIV-1 NL4-3ΔenvLuc VSVg virions or transfected with a plasmid encoding the HIV-1 NL4-3ΔenvLuc genome, and luciferase activity was measured 48 h later (Fig. 3A). As a control, luciferase-targeted siRNAs inhibited HIV-1 expression in both conditions. Similarly, decreasing expression of cyclin T1, a factor required for the transcriptional elongation of HIV-1 genes, severely affected HIV-1 expression from both transfected and integrated genome. In contrast, VBP1 knockdown had no effect on HIV-1 expression from the transfected plasmid pNL4-3ΔenvLuc (Fig. 3A Right), whereas viral expression was decreased 10-fold upon infection of cells treated with VBP1-targeted siRNAs (Fig. 3A Left). These results thus show that VBP1 is not involved in HIV gene expression when the integration step is by-passed by direct transient transfection of the viral genome.
Fig. 3.
Integration is required before transcriptional control of HIV by VBP1. (A) HeLa cells were transfected with indicated siRNAs and subsequently infected with NL4-3ΔenvLuc VSVg virus (Left) or transfected with a plasmid encoding the NL4-3ΔenvLuc genome (Right). Luciferase activity was monitored in cell lysates 48 h after infection or transfection. (B) HeLa LTR-Luc cells were first transfected with indicated siRNAs and subsequently transfected with the indicated amount of a plasmid encoding Tat. Luciferase activity was monitored in cell lysates 48 h after transfection.
To test whether the effect of VBP1 depends on integrase-dependent or -independent integration, we used distinct clones stably transfected with luciferase gene under the control of HIV-1 promoter and measured the Tat-induced transcription of luciferase gene (36). Luciferase or cyclin T1-targeted siRNAs strongly inhibited luciferase expression, whereas silencing VBP1 but also pVHL in these cells had no major effect on Tat-mediated transactivation of the integrated HIV-1 promoter per se (Fig. 3B and data not shown for other independent clones). Together, these data indicate that VBP1 and Cul2/VHL do not directly interfere with the transcription machinery but are required for HIV1 gene expression when the viral genome had been integrated through an integrase-dependent pathway.
VBP1-Containing Prefoldin and VHL Ubiquitin Ligase Are Involved in Integrase Ubiquitylation and Degradation.
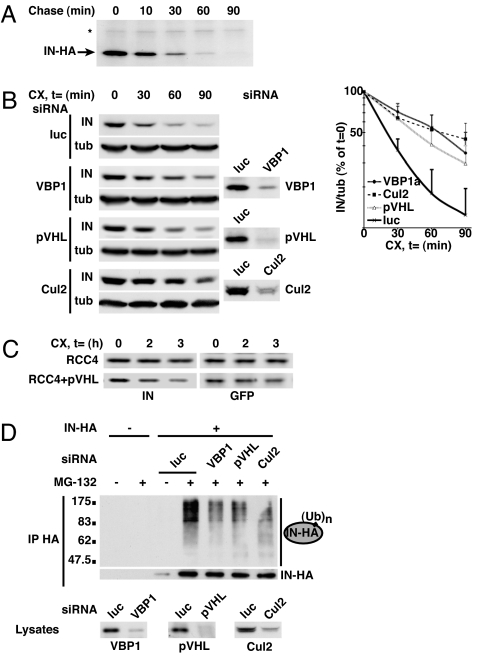
The well known involvement of the Cul2/VHL ubiquitin ligase complex in the ubiquitin-mediated degradation of cellular targets, particularly the α-subunits of the hypoxia-inducible transcription factor (HIF), led us to test whether VBP1 and Cul2/VHL control IN degradation. Pulse–chase assay showed that IN-HA was a very unstable protein with an estimated half-life of 11 min (Fig. 4A). Similar results were obtained by treatment of IN-HA cells with cycloheximide that led to a 23-min half-life (Fig. 4B). In addition, a 3-h treatment with the proteasome inhibitor MG-132 resulted in the 40-fold accumulation of IN-HA protein levels. It also led to the accumulation of ubiquitylated forms of IN (Fig. 4D). These results are consistent with previous reports showing that proteasome inhibition leads to an inhibition of IN degradation (24, 28–30). Mutational analysis of lysine residues indicated that replacing lysine 211, 215, 219, or 273 with arginines slowed down degradation of IN by a factor of 3 (data not shown), thus suggesting that these residues represent the major targets for ubiquitin-mediated degradation.
Fig. 4.
Prefoldin and Cul2/VHL complexes are involved in the ubiquitin–proteasome-dependent degradation of IN. (A) IN-HA cells were pulse-labeled with [35S]methionine/cysteine and subsequently chased for indicated time periods prior to immunoprecipitation by using anti-HA antibody. Immunoprecipitated proteins were analyzed by SDS/PAGE and fluorography. IN-HA and a stable contaminating protein (*) reflecting loading are visualized. Quantifications of two independent experiments resulted in an estimated IN half-life of 11 min. (B) IN-HA cells were transfected with siRNA specifically directed against VBP1 (VBP1a), pVHL, or Cul2 or with control luciferase (luc)-directed siRNA. Cells were subsequently treated with the protein synthesis inhibitor cycloheximide (100 μg/ml) for the indicated periods of time prior to lysis, and analyzed by Western blotting with anti-HA antibody and anti-α-tubulin antibody as an internal control. The effect of siRNAs on protein expression was monitored with specific antibodies. Chemiluminescence of the blots was acquired with a Fuji CCD camera (Kanagawa, Japan). For each condition, the IN-HA chemiluminescence signal was quantified by using Image Gauge software and normalized to the α-tubulin signal. Results from five to seven independent experiments are represented on the right. (C) pVHL-negative RCC4 cells stably transfected with pVHL (RCC4+pVHL) or not (RCC4) were transiently cotransfected with IN-HA and GFP expression plasmids. Cells were subsequently treated with cycloheximide (100 μg/ml) for the indicated times prior to lysis. Equal amounts of total protein lysates were then analyzed by Western blotting with anti-HA and anti-GFP antibodies. Chemiluminescence of the blots was quantified as in B, and the IN-HA signal was normalized to the GFP transfection control signal. (D) IN-HA or HeLa cells were transfected indicated siRNAs and subsequently treated with the proteasome inhibitor MG-132 (20 μM) or DMSO prior to lysis. Equal amounts of total cellular proteins were immunoprecipitated by using anti-HA antibody. Immunoprecipitated proteins were analyzed by Western blotting with anti-HA or anti-ubiquitin antibody (IP HA). VBP1 expression in cell lysates was monitored by anti-VBP1 immunoblotting (lysates).
To determine whether VBP1 and Cul2/VHL control IN turnover, IN-HA stability was analyzed in cells treated with VBP1 or Cul2/VHL complex-directed siRNAs. Representative data from five to seven independent experiments revealed that knockdown of VBP1, pVHL, or Cul2 expression by specific siRNA resulted in a slower IN degradation compared with control cells treated with a control siRNA. Notably, IN half-life increased from 23 min in control cells to 65, 49, and 62 min in VBP1, pVHL, and Cul2 siRNA-treated cells, respectively (Fig. 4B). IN was also stabilized, although to a lower extent, upon knockdown of prefoldin 5 and CCT2 [supporting information (SI) Fig. 5].
To confirm the role of pVHL in IN degradation, IN turnover was analyzed in the renal cell carcinoma cell line RCC4, which is deficient in pVHL expression. For this purpose, an IN-HA expression plasmid was transiently transfected into RCC4 cells together with a GFP-encoding vector as an internal control for transfection, and IN degradation was monitored by Western blotting after cycloheximide cell treatment. Quantification of GFP and IN protein levels showed that IN was stabilized in RCC4 cells, even after translation inhibition with cycloheximide (Fig. 4C). In contrast, expression of pVHL in RCC4 cells partially restored IN degradation (Fig. 4C) with a 36% decrease in the IN protein level after a 3-h treatment with cycloheximide. Together, these results indicate that IN stabilization in RCC4 cells results from an altered Cul2/VHL-mediated degradation pathway, at least to some degree.
To further characterize the role of VBP1 and Cul2/VHL in IN turnover, their effect on IN ubiquitylation was specifically analyzed. Notably, cell treatment with the proteasome inhibitor MG-132 led to the accumulation of polyubiquitylated IN, indicating that polyubiquitylation of IN precedes its degradation by the proteasome (Fig. 4D). Silencing VBP1 or Cul2/VHL resulted in a 40–55% reproducible decrease in the accumulation of polyubiquitylated forms of IN after proteasome inhibition (Fig. 4D). These data thus clearly show not only that both VBP1 and the Cul2/VHL ligase complex participate in the ubiquitylation of IN before its proteasome-mediated degradation but also strongly suggest that Cul2/VHL is a key ubiquitin ligase responsible for IN ubiquitylation and that VBP1 may allow the prefoldin chaperone to target IN to the Cul2/VHL ligase. Finally, we mutated target lysine residues for ubiquitin-mediated degradation in the context of the full-length virus and analyzed the infectivity of the resulting virus. Although lysine 273 is shared with the HIV-1 vif protein and cannot be mutated in the viral context, mutation of lysines 211, 215, and 219 into arginine already led to a 60% decrease of viral infectivity (SI Fig. 6), a result that corroborates a role for ubiquitin-mediated degradation in HIV-1 replication.
Taken together, these results support the notion that VBP1 and Cul2/VHL strictly act at a postintegration step of HIV-1 replication by regulating the level of transcription of integrated provirus. However, VBP1 is not required for HIV transcription when the integrase-dependent integration step is by-passed by direct transfection of the viral genome. Because (i) HIV-1 integrase catalyzes integration of the viral genome into the host genome, (ii) VBP1 (and VHL) does not affect integrase activity but controls integrase stability, and (iii) no viral target other than integrase has been found for VBP1, VBP1/VHL-mediated IN degradation is likely required for the proper transcription of viral genes.
Discussion
HIV-1 IN has been reported to be degraded by the ubiquitin–proteasome system in an N-end-rule-dependent and -independent pathway (24, 28–31). Here, we show that the prefoldin chaperone subunit VBP1 specifically interacts with IN and mediates IN interaction with pVHL, a substrate-specific adaptor of the Cul2-based VHL ubiquitin ligase. The prefoldin chaperone and the Cul2/VHL ligase mediate IN polyubiquitylation that leads to subsequent proteasome-dependent degradation. By delivering IN to Cul2/VHL, the prefoldin chaperone would thus target its protein substrate IN for ubiquitin–proteasome degradation, thus contrasting with its well described function in protein folding. Previously, prefoldin has been reported to mediate folding of protein substrates such as actin and tubulin by specifically targeting these substrates to the CCT chaperonin (34, 35, 37). Our data now suggests that prefoldin plays a pivotal role in a “folding versus degradation” checkpoint by cooperating with both other chaperones and the ubiquitin–proteasome system. Respective roles of prefoldin and Cul2/VHL ligase in IN ubiquitylation can be compared with the functions of Hsp70 or Hsp90 chaperones and CHIP ubiquitin ligase in sorting specific substrates to the proteasome. Besides their role in protein folding, Hsp70 and Hsp90 chaperones have been reported to trigger polyubiquitylation and proteasomal degradation of glucocorticoid receptor (GR) or cystic fibrosis transmembrane conductance regulator (CFTR) by directly recruiting the CHIP E3 ligase (38). Data presented here thus provide another illustration of the tight link between protein folding and protein degradation machineries.
IN was previously reported to be protected from proteasome-mediated degradation by two cellular binding partners, LEDGF/p75 and hRad18 (20, 29). Interestingly, the VBP1-binding domain of IN as defined here (between residues 43 and 195) includes the p75-interacting domain (24, 39). Moreover, screening a two-hybrid library of HIV-1 IN random mutants by using LEDGF/p75 or VBP1 as a bait led to the identification of different mutations in the IN catalytic core domain, impairing its interaction with both LEDGF/p75 and VBP1 (S.E., J.-C.R., and R.B., unpublished data). These data are thus consistent with an overlapping binding domain of both cellular proteins within IN. LEDGF/p75 may indeed protect IN from proteasome degradation by masking its interaction site with VBP1. It is worth noting that VBP1 knockdown does not affect the intracellular distribution of IN or its tethering to mitotic chromosomes (data not shown), suggesting that interaction with VBP1 might occur after IN has been targeted to chromatin in a LEDGF/p75-dependent manner.
Analysis of the effect of VBP1 and pVHL knockdown upon cell infection with NL4-3ΔenvLuc VSVg virions clearly indicates that both cellular proteins participate in HIV-1 gene expression. Interestingly, we found that reduction of VBP1 expression by RNAi specifically inhibited viral transcription without significantly affecting the amount of reverse-transcribed viral DNA or integrated proviral DNA. Although we cannot definitely exclude a role of VBP1 in regulating transcription by recruiting transcriptional coactivators to the integrated HIV promoter, no effect of VBP1 knockdown could be observed on HIV-1 promoter-driven transcription when the integrase-dependent integration step is by-passed by direct-transient or stable transfection. These data, therefore, strongly suggest that VBP1 and VHL are required for the proper transition between integration and transcription of the viral genome. In addition, screening a highly complex library of HIV-1 random fragments by using VBP1 as bait led exclusively to the IN gene indicating that IN is the unique target of VBP1 within proteins encoded by the HIV-1 viral genome. Together, these data support a role for VBP1 in HIV-1 replication, mediated by its interaction with IN, and strongly suggest that IN degradation by the prefoldin–VHL–proteasome pathway would play an important role for efficient transcription of viral genes after IN-catalyzed integration of the proviral DNA into the host genome is accomplished.
IN degradation may be necessary for the correct repair of the integration intermediate by cellular enzymes and consequently for viral transcription. Requirement for an active IN disassembly from the strand-transfer products prior to gap repair has already been suggested by in vitro studies (11, 17). Such a requirement of IN degradation prior to repair is likely to be unique. Rearrangement of the Ig and T cell receptor genes is initiated by the IN-related recombinase RAG1/2, which introduces dsDNA breaks at recombination signal sequences that are subsequently joined by the cellular nonhomologous DNA end-joining (NHEJ) machinery. After cleavage, the RAG1/2 recombinase remains tightly bound to the recombination signal sequence ends and sequesters them from the repair machinery (40, 41). Specific remodeling or disassembly of this complex has been proposed to allow the joining to proceed (42). Furthermore, whereas RAG1 autoubiquitylation has been suggested to assist remodeling of the postcleavage complex (43), RAG2 has been found to undergo Skp2-SCF-mediated ubiquitylation and degradation (44), thereby suggesting that RAG1/2 ubiquitylation might promote joining of the cleaved recombination signal sequence.
A role for remodeling in transposition/integration processes catalyzed by the polynucleotidyl transferase family of enzymes is also illustrated during transposition of the Escherichia coli phage Mu. This process is catalyzed by the bacteriophage-encoded MuA transposase that remains tightly bound to the strand-transfer product at the Mu DNA ends after the strand-transfer reaction, thereby inhibiting assembly of the bacterial DNA-replication machinery and lytic growth (45). The recombination–replication transition of the Mu life cycle requires destabilization of the MuA–DNA complex by the bacterial chaperone molecule ClpX, which unfolds and releases a subset of MuA subunits from the strand-transfer complex, thus allowing recruitment of the replication machinery (46–49). Similarly to Mu transposition, IN disassembly from the proviral DNA ends by the prefoldin–VHL–proteasome machinery after HIV-1 integration could be required for viral transcription to proceed. Furthermore, a protective effect mediated by LEDGF/p75 or hRad18 against IN degradation could be compared with a function of the phage transposition activator MuB that prevents MuA remodeling during the recombination process through a MuA binding site overlapping ClpX binding sequence (50).
In conclusion, findings reported here support the notion that the regulation of HIV-1 IN stability plays a major role at specific and crucial steps of the viral replication cycle. A thorough understanding of the consequences of the prefoldin-VHL-mediated IN degradation on remodeling of the strand-transfer complex is likely to provide insights into the integration–transcription transition of the viral life cycle.
Methods
Two-hybrid screenings using HIV-1 integrase as bait were performed as described in ref. 24.
To create the HA epitope-tagged IN-expression construct pcDNA3-INsalaHA, the FLAG epitope of the pCEP-INsalaFLAG construct (18) was replaced by the HA epitope (GYPYDVPDYA).
Plasmid constructions, primers, siRNA, and conditions for cell culture and transfection and infection assays are detailed in SI Text.
Experimental procedures that were used for the analysis of integrase ubiquitylation and degradation, as well as for the quantification of viral DNA and RNA, are precisely described in SI Text.
Supplementary Material
Acknowledgments
We thank Wilhelm Krek (Institute of Cell Biology, Zurich, Switzerland) for the gift of the pcDNA3-HA-pVHL plasmid and the anti-pVHLCT antibody, Jacques Pouysségur (Centre Antoine Lacassagne, Nice, France) for the gift of the RCC4 and RCC4+pVHL cells, Zeger Debyser (Katholieke Universiteit, Leuven, Belgium) for the gift of the pCEP-INsalaFLAG plasmid, Jean-François Mouscadet and Gilles Divita for critical reading of the manuscript, and the members of the C.D. laboratory for helpful discussions. This work was supported by grants from Agence Nationale de Recherches sur le Sida (to S.E., R.B., and C.D.) and Sidaction (to R.B.), European Commission “Hidden HIV Challenge” Grant FP6–2003-LIFESCIHEALTH-3/012182 (to S.E.), and “Targeting Replication and Integration of HIV” Grant LSHB-CT-2003-503480 (to R.B.). A.M. was supported by a postdoctoral fellowship from Agence Nationale de Recherches sur le Sida, and N.K. was supported by a postdoctoral fellowship from the European Commission project “Hidden HIV Challenge.”
Abbreviations
- VHL
von Hippel–Lindau
- VBP1
von Hippel–Lindau binding protein 1
- pVHL
VHL protein
- IN
integrase
- IN-HA
HA-tagged IN
- Cul2
cullin2
- LEDGF/p75
lens epithelium-derived growth factor/transcription coactivator p75
- NL4-3ΔenvLuc VSVg
NL4-3Δenv virus pseudotyped with VSV-G envelope and containing the luciferase gene inserted into the nef gene.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705162104/DC1.
References
- 1.Brown PO. Curr Top Microbiol Immunol. 1990;157:19–48. doi: 10.1007/978-3-642-75218-6_2. [DOI] [PubMed] [Google Scholar]
- 2.Hehl EA, Joshi P, Kalpana GV, Prasad VR. J Virol. 2004;78:5056–5067. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tasara T, Maga G, Hottiger MO, Hubscher U. FEBS Lett. 2001;507:39–44. doi: 10.1016/s0014-5793(01)02945-3. [DOI] [PubMed] [Google Scholar]
- 4.Tsurutani N, Kubo M, Maeda Y, Ohashi T, Yamamoto N, Kannagi M, Masuda T. J Virol. 2000;74:4795–4806. doi: 10.1128/jvi.74.10.4795-4806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. J Virol. 1999;73:2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu K, Dobard C, Chow SA. J Virol. 2004;78:5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouyac-Bertoia M, Dvorin JD, Fouchier RA, Jenkins Y, Meyer BE, Wu LI, Emerman M, Malim MH. Mol Cell. 2001;7:1025–1035. doi: 10.1016/s1097-2765(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 8.Gallay P, Hope T, Chin D, Trono D. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda T, Nishitsuji H, Zhou X, Nara N, Ohashi T, Kannagi M, Masuda T. J Virol. 2004;78:11563–11573. doi: 10.1128/JVI.78.21.11563-11573.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukovsky A, Gottlinger H. J Virol. 1996;70:6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz RA, Skalka AM. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 13.Engelman A, Mizuuchi K, Craigie R. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 14.Brown PO, Bowerman B, Varmus HE, Bishop JM. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison V, Abrams H, Roe T, Lifson J, Brown P. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara T, Mizuuchi K. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 17.Yoder KE, Bushman FD. J Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 19.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 20.Mulder LC, Chakrabarti LA, Muesing MA. J Biol Chem. 2002;277:27489–27493. doi: 10.1074/jbc.M203061200. [DOI] [PubMed] [Google Scholar]
- 21.Parissi V, Calmels C, De Soultrait VR, Caumont A, Fournier M, Chaignepain S, Litvak S. J Virol. 2001;75:11344–11353. doi: 10.1128/JVI.75.23.11344-11353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Violot S, Hong SS, Rakotobe D, Petit C, Gay B, Moreau K, Billaud G, Priet S, Sire J, Schwartz O, et al. J Virol. 2003;77:12507–12522. doi: 10.1128/JVI.77.23.12507-12522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 24.Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, et al. J Biol Chem. 2005;280:25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 25.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 26.Jacque JM, Stevenson M. Nature. 2006;441:641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- 27.Turlure F, Devroe E, Silver PA, Engelman A. Front Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 28.Devroe E, Engelman A, Silver PA. J Cell Sci. 2003;116:4401–4408. doi: 10.1242/jcs.00747. [DOI] [PubMed] [Google Scholar]
- 29.Llano M, Delgado S, Vanegas M, Poeschla EM. J Biol Chem. 2004;279:55570–55577. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- 30.Mulder LC, Muesing MA. J Biol Chem. 2000;275:29749–29753. doi: 10.1074/jbc.M004670200. [DOI] [PubMed] [Google Scholar]
- 31.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuchiya H, Iseda T, Hino O. Cancer Res. 1996;56:2881–2885. [PubMed] [Google Scholar]
- 33.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 35.Geissler S, Siegers K, Schiebel E. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S. EMBO J. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU. EMBO J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esser C, Alberti S, Hohfeld J. Biochim Biophys Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 39.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Proc Natl Acad Sci USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agrawal A, Schatz DG. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 41.Jones JM, Gellert M. Proc Natl Acad Sci USA. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramsden DA, Gellert M. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 43.Jones JM, Gellert M. Proc Natl Acad Sci USA. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Desiderio S. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 45.Nakai H, Kruklitis R. J Biol Chem. 1995;270:19591–19598. doi: 10.1074/jbc.270.33.19591. [DOI] [PubMed] [Google Scholar]
- 46.Levchenko I, Luo L, Baker TA. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 47.Kruklitis R, Welty DJ, Nakai H. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 48.Burton BM, Williams TL, Baker TA. Mol Cell. 2001;8:449–454. doi: 10.1016/s1097-2765(01)00307-0. [DOI] [PubMed] [Google Scholar]
- 49.Burton BM, Baker TA. Chem Biol. 2003;10:463–472. doi: 10.1016/s1074-5521(03)00102-9. [DOI] [PubMed] [Google Scholar]
- 50.Levchenko I, Yamauchi M, Baker TA. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.