Abstract
Intron-containing pre-mRNAs are normally retained in the nucleus until they are spliced to produce mature mRNAs that are exported to the cytoplasm. Although the detailed mechanism is not well understood, the formation of splicing-related complexes on pre-mRNAs is thought to be responsible for the nuclear retention. Therefore, pre-mRNAs containing suboptimal splice sites should tend to leak out to the cytoplasm. Such pre-mRNAs often contain purine-rich exonic splicing enhancers (ESEs) that stimulate splicing of the adjacent intron. Here, we show that ESEs per se possess an activity to retain RNAs in the nucleus through a saturable nuclear retention factor. Cross-competition experiments revealed that intron-containing pre-mRNAs (without ESEs) used the same saturable nuclear retention factor as ESEs. Interestingly, although intronless mRNAs containing ESEs were also poorly exported, spliced mRNAs produced from ESE-containing pre-mRNAs were efficiently exported to the cytoplasm. Thus, the splicing reaction can reset the nuclear retention state caused by ESEs, allowing nuclear export of mature mRNAs. Our results reveal a novel aspect of ESE activity that should contribute to gene expression and RNA quality control.
Keywords: nuclear export, quality control
The nuclear envelope separates eukaryotic cells into two major compartments, the nucleus and the cytoplasm. This compartmentalization necessitates transport through the nuclear pore complexes, the channels for material exchange across the nuclear envelope barrier (reviewed in ref. 1). The vast majority of RNA species, after their synthesis and processing in the nucleus, are exported to the cytoplasm. Some specific RNA species, however, are not exported but instead stay in the nucleus. Such RNAs include U6 small nuclear RNAs (snRNAs), small nucleolar RNAs, small Cajal body RNAs, some mRNA-like noncoding RNAs, inosine-containing RNAs, and various unprocessed immature RNAs (reviewed in refs. 2–6). Among them, intron-containing pre-mRNAs are retained in the nucleus as a part of the RNA quality-control mechanism that ensures appearance in the cytoplasm of only mature mRNAs. This retention is thought to be achieved as a result of the formation of splicing-related complexes, although the detailed mechanism is not well understood (reviewed in refs. 6 and 7).
Properly processed RNAs are generally exported to the cytoplasm, but different RNA species, such as tRNAs, U snRNAs, mRNAs, and rRNAs, use distinct export pathways, i.e., distinct sets of export factors (reviewed in ref. 8). Accumulating evidence shows that the pathway of RNA export can influence the fate of a given RNA in the cytoplasm (8), indicating the biological importance of the choice of RNA export pathway. This means that the cellular export machinery must be able to discriminate distinct RNA species, and therefore each RNA species should have its own distinguishing features.
We have been searching for the distinguishing features of mRNA, termed mRNA ID elements. We previously found that the presence of introns in mRNA precursors is one such feature. If a pre-mRNA intron was artificially inserted into U1 snRNA, the spliced U1 snRNA was exported via the mRNA export pathway instead of the U snRNA pathway, indicating that introns can function as an identity element for mRNA export (9). We subsequently found that the presence of a stretch of unstructured RNA region of certain length can function as a splicing-independent mRNA ID element (10).
In the course of attempts to search for more such ID elements, we hypothesized, for the following reasons, that purine-rich exonic splicing enhancers (ESEs) may function as such an ID element of mRNA. The ESE was originally found in the IgM and cardiac troponin T genes and subsequently in many other genes (reviewed in refs. 11–13). ESEs are purine-rich RNA sequences in exons that stimulate the splicing of the adjacent introns with suboptimal splice sites. Artificial purine-rich repeat sequences such as (GAA)n exert strong enhancer activity (14, 15).
ESEs are recognized and bound by RS domain-containing proteins, such as SF2/ASF, Tra2β, and SRp30c (15–17), and these SR proteins, in turn, increase the efficiency of recruitment of U2AF to the 3′ splice site, leading to the recruitment of U2 snRNP to the branch point (11–13). ESEs and bound factors can also stimulate the 5′ splice site recognition and may also be involved in the interaction between 5′ and 3′ splicing complexes across introns and/or exons (12, 18–20). On the other hand, some shuttling SR proteins, such as SF2/ASF, SRp20, and 9G8, can interact with mRNA export factor TAP/p15 heterodimer and recruit the factor onto RNA (21–24). Therefore, we hypothesized that ESEs may be able to induce mRNA export through the recruitment of SR proteins and TAP/p15.
To test this hypothesis, various ESE sequences were inserted into U1 RNA, and the export of the resultant artificial RNAs was examined in Xenopus oocyte microinjection experiments to determine whether ESEs could function as an mRNA ID element. Contrary to our expectation, it turned out that ESEs function as an RNA nuclear retention element rather than an mRNA ID element, as will be described.
Results
ESE-Containing U1 snRNAs Are Poorly Exported.
To examine whether ESEs can function as an mRNA ID element, one or multiple copies of purine-rich ESE sequences from various genes were inserted into U1ΔSm RNA, and export of the resultant chimeric U1 RNAs was examined in microinjection experiments using Xenopus oocytes (Fig. 1). If these natural ESEs function as we originally anticipated, they should be able to shift export of U1-ESE RNAs from the U snRNA pathway to the mRNA pathway, as was the case with other mRNA ID elements.
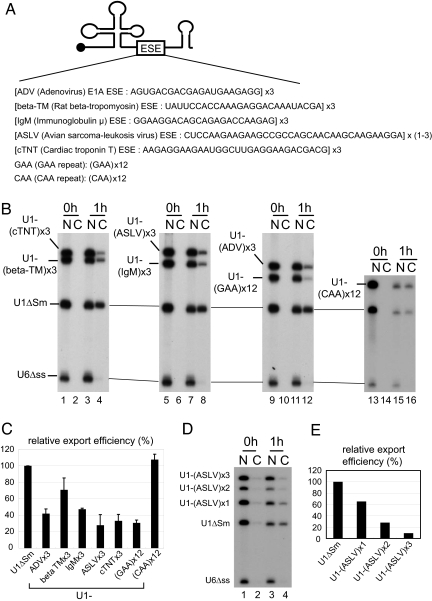
Fig. 1.
Effect of ESEs on U1 RNA export. (A) Schematic representation of U1-ESE derivatives used for the export analysis. One or multiple copies of purine-rich ESE sequences from various genes were inserted into U1ΔSm RNA. (B) A mixture of in vitro-transcribed 32P-labeled RNAs containing U1-ESE, U1ΔSm, and U6Δss RNAs was injected into the nucleus of Xenopus oocytes. U6Δss RNA was uncapped, and all of the other RNAs were m7G-capped. RNA was extracted from nuclear (N) and cytoplasmic (C) fractions immediately (0 h; lanes 1, 2, 5, 6, 9, 10, 13, and 14) or 1 h (1 h; lanes 3, 4, 7, 8, 11, 12, 15, and 16) after injection and analyzed by 8% denaturing PAGE. (C) Quantitation of RNA export from three independent experiments as in B. Averages and standard deviations are shown. (D) The same as in B except that one to three copies of ASLV ESE were used. (E) Quantitation of D.
A mixture of in vitro-transcribed32P-labeled RNAs containing U1-ESE RNAs, U1ΔSm RNA, and nonexported U6Δss control RNA was injected into the nucleus of Xenopus oocytes (Fig. 1 B–E). U6Δss RNA was uncapped, and all of the other RNAs were m7G-capped. Immediately after nuclear injection, all of the RNAs were nuclear (Fig. 1B, 0h). After 1 h, U1ΔSm RNA was efficiently exported, whereas U6ΔssRNA stayed in the nucleus as expected (Fig. 1B, 1h). However, U1-ESE RNAs were exported poorly as compared with the parental U1ΔSm RNA (Fig. 1B; Fig. 1C for quantitation). In addition, an artificial GAA repeat sequence that had been shown to exert a strong splicing enhancer activity (14, 15) reduced U1 RNA export, whereas the control CAA repeat did not have such activity (Fig. 1 B and C). The strength of export inhibitory activity of a given ESE appeared to be well correlated with the strength of its splicing enhancing activity. Among the natural ESEs tested here, avian sarcoma-leukosis virus (ASLV) and cardiac troponin T (cTNT) ESEs showed the strongest activity of both splicing enhancement and nuclear retention, whereas β-tropmyosin (β-TM) ESE showed the weakest activity of both (see ref. 14 and Fig. 1C). Moreover, only one copy of ASLV ESE significantly inhibited U1 RNA export, and more copies exerted greater inhibition (Fig. 1 D and E). These results showed that ESE sequences have export inhibitory activity rather than the mRNA export-inducing activity that was initially expected.
Nevertheless, it was possible that the observed inefficient export of U1-ESE RNAs used the mRNA export pathway. To test this issue, we examined the export pathway of U1-(GAA)12 RNA by the use of CRM1-specific export inhibitor BSA-NES [see supporting information (SI) Fig. 6]. The inefficient export of U1-(GAA)12 RNA, and the export of U1ΔSm RNA, was completely inhibited by BSA-NES, whereas the control conjugate BSA-mut hardly affected RNA export, suggesting that the inefficient export used the U snRNA pathway rather than the mRNA pathway. Therefore, it is very unlikely that ESEs per se can function as mRNA ID elements.
ESEs Retain RNA in the Nucleus Through a Saturable Retention Factor.
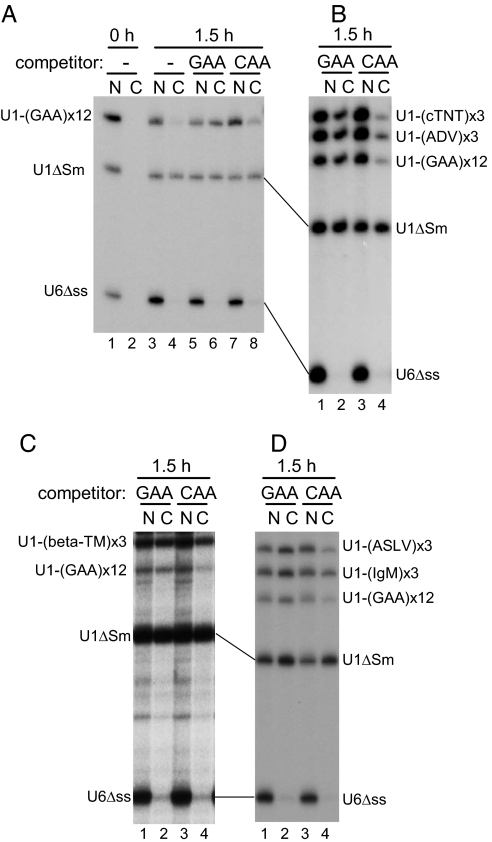
To obtain a clue regarding how ESEs inhibit RNA export, a cross-competition experiment was performed (Fig. 2). Various 32P-labeled U1-ESE RNAs were injected into the nucleus of Xenopus oocytes as in Fig. 1, together with excess unlabeled uncapped U1-(GAA)12 RNA competitor (100 fmol per oocyte; 20- to 30-fold excess over labeled RNAs) or with the same amount of a control competitor, U1-(CAA)12 RNA, or without competitors, and RNA export was examined (Fig. 2). All of the labeled U1-ESE RNAs were more or less poorly exported in the absence of the competitors (Fig. 2A, lanes 1–4) and the presence of the control competitor U1-(CAA)12 RNA (Fig. 2A, lanes 7 and 8; Fig. 2B, lanes 3 and 4; Fig. 2C, lanes 3 and 4; and Fig. 2D, lanes 3 and 4). However, in the presence of U1-(GAA)12 RNA competitor, the export of all of the labeled U1-ESE RNAs was significantly stimulated, whereas neither the export of U1ΔSm RNA nor the nuclear retention of U6Δss RNA was affected (Fig. 2A, lanes 5 and 6; Fig. 2B, lanes 1 and 2; Fig. 2C, lanes 1 and 2; and Fig. 2D, lanes 1 and 2), indicating that the nuclear retention activity of various ESEs could be blocked by competition by the common competitor U1-(GAA)12 RNA. This result can be interpreted to mean that a common saturable nuclear retention factor is involved for all of the tested ESEs.
Fig. 2.
Cross-competition experiments with U1-ESE RNAs. The same as in Fig. 1B except that the RNA mixture was injected either alone (A, lanes 1–4) or with 100 fmol per oocyte of unlabeled, uncapped U1-(GAA)12 (GAA; A, lanes 5 and 6; B–D, lanes 1 and 2) or U1-(CAA)12 (CAA; A, lanes 7 and 8; B–D, lanes 3 and 4) competitor RNA. RNA was analyzed immediately (0 h; A, lanes 1 and 2) or 1 h (1.5 h; lanes 3–8) after injection. N, nuclear fractions; C, cytoplasmic fractions.
Pre-mRNAs Use the Same Saturable Nuclear Retention Factor as ESEs.
To examine whether regular pre-mRNAs without ESEs use the same nuclear retention mechanism as RNAs with ESEs, the following cross-competition experiments were performed. 32P-labeled ftz pre-mRNA without ESEs (pre-ftz) was injected into the nucleus of Xenopus oocytes, together with various 32P-labeled reference RNAs, including dihydrofolate reductase (DHFR) mRNA, U1-(GAA)12 RNA, U1ΔSm RNA, U6Δss RNA, and tRNAphe (Fig. 3B). Under normal conditions, pre-ftz was spliced efficiently almost to completion, and the spliced ftz RNA, as well as DHFR mRNA, U1ΔSm RNA, and tRNAphe, was efficiently exported to the cytoplasm (Fig. 3B, lanes 1–4). Only a small amount of pre-ftz was seen in the cytoplasm (Fig. 3B, lane 4). To investigate the behavior of pre-ftz in detail, splicing was inhibited by preinjection of an antisense oligo nucleotide against U2 snRNA, which causes effective U2 snRNA depletion by the endogenous RNaseH activity (9). Depletion of U2 but not U1 was confirmed by Northern blot analysis (Fig. 3A). Upon such depletion of U2, the production of spliced ftz RNA was severely inhibited and pre-ftz RNA accumulated (Fig. 3B, lanes 5–10). Both pre-ftz and U1-(GAA)12 were still retained in the nucleus (8.1% and 25% in the cytoplasm, respectively; Fig. 3B, lanes 5 and 6), suggesting that U2 snRNP per se is not the retention factor. However, when a saturating amount of unlabeled U1-(GAA)12 RNA competitor was coinjected, export of pre-ftz and U1-(GAA)12 was induced (32% and 53% in the cytoplasm, respectively; Fig. 3B, lanes 7 and 8), whereas the same amount of control competitor U1-(CAA)12 had no effect on these RNAs (Fig. 3B, lanes 9 and 10). The export of DHFR mRNA was weakly inhibited by both competitors for some unknown reason. Taken together, these results indicate that the nuclear retention of pre-mRNAs shares a common factor with the nuclear retention of ESEs and that the common retention factor is not likely to be U2 snRNP itself.
Fig. 3.
Effect of U1-(GAA)12 competitor on the nuclear retention of pre-ftz. (A) An antisense oligo against U2 snRNA was injected into the cytoplasm of Xenopus oocytes to destroy endogenous U2 RNA (ΔU2). Water was injected as a control (−). After 20 h of incubation, RNA was recovered, and the integrity of endogenous U1 and U2 snRNAs was examined by Northern blot analysis. (B) A mixture of 32P-labeled ftz pre-mRNA without ESE (pre-ftz), DHFR mRNA, U1-(GAA)12 RNA, U1ΔSm RNA, U6Δss RNA, and tRNAphe was injected into the nucleus of control (lanes 1–4) or ΔU2 (lanes 5–10) oocytes, either alone (lanes 1–6) or with GAA (lanes 7 and 8) or CAA (lanes 9 and 10) competitor as in Fig. 2. RNA was analyzed immediately (0 h; lanes 1 and 2) or 2 h (2 h; lanes 3–10) after injection. N, nuclear fractions; C, cytoplasmic fractions.
Splicing Can Reset the Nuclear Retention State Caused by ESEs.
Because ESEs enhance splicing and thereby increase gene expression from genes containing weak splice sites, it would not make sense for ESEs to inhibit the export of the spliced RNAs. To obtain a clue regarding this inconsistency, the effect of ESEs on mRNA export rather than U snRNA export was examined (Fig. 4).
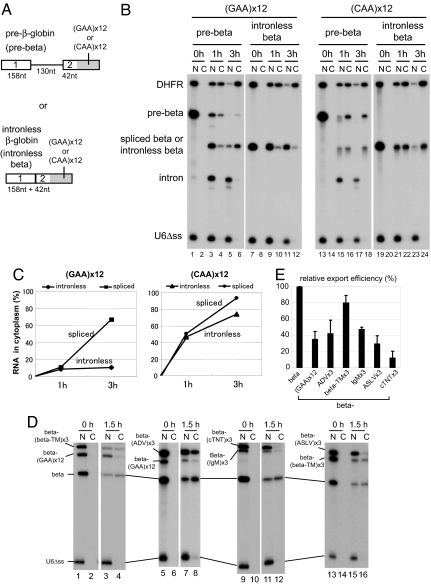
Fig. 4.
Effect of splicing on the nuclear retention by ESEs. (A) Schematic representation of β-globin derivatives. GAA or CAA repeat sequence was fused to either β-globin pre-mRNA (pre-beta; Upper) or intronless β-globin mRNA (intronless beta; Lower). The sizes of the exons and an intron are indicated. (B) 32P-labeled β-globin pre-mRNA (pre-beta) or intronless β-globin mRNA (intronless beta), fused to either GAA (lanes 1–12) or CAA (lanes 13–24) repeat sequence, was injected into the nucleus of Xenopus oocytes, together with 32P-labeled DHFR mRNA and U6Δss RNA. RNA was analyzed immediately (0 h; lanes 1, 2, 7, 8, 13, 14, 19, and 20), 1 h (1 h; lanes 3, 4, 9, 10, 15, 16, 21, and 22), or 3 h (3 h; lanes 5, 6, 11, 12, 17, 18, 23, and 24) after injection. (C) Export kinetics of intronless beta or spliced beta RNA derivatives from B. (D) The same as in Fig. 1 except that an intronless β-globin mRNA instead of U1 was fused to various ESEs and the incubation was 0 or 1.5 h. (E) Quantitation of RNA export from three independent experiments as in D. N, nuclear fractions; C, cytoplasmic fractions.
The GAA or CAA repeat sequence was fused to either an intronless mRNA derived from β-globin cDNA (Fig. 4A Lower) or intron-containing β-globin pre-mRNA (Fig. 4A Upper), and RNA export was examined (Fig. 4 B and C). When the CAA repeat was used, both the intronless β-mRNA and the spliced mRNA produced from pre-β were exported efficiently with similar kinetics in this condition (Fig. 4B and C Right).
When the intronless β-mRNA with the GAA repeat was used, however, its export was severely inhibited (Fig. 4B, lanes 7–12 and Fig. 4C Left). This was also the case with various natural ESEs (Fig. 4 D and E for quantitation). In contrast, when pre-β with the GAA repeat was used, the spliced mRNA was exported quite efficiently despite the fact that the spliced mRNA contained the GAA repeat (Fig. 4B, lanes 1–6 and C Left). These results indicated that pre-mRNA splicing can reset the nuclear retention state caused by ESEs.
Although pre-β-RNA is already an efficient splicing substrate, small enhancement of splicing was observed in the presence of the GAA repeat, as judged by the amount of excised intron lariat (Fig. 4B, compare lanes 3 and 5 with 15 and 17). The excised intron was exclusively seen in the nucleus. Furthermore, the amount of pre-mRNA that leaked out to the cytoplasm was lower with the GAA repeat as compared with the CAA repeat (Fig. 4B, compare lanes 3–6 with 15–18), suggesting that the GAA repeat had an activity preventing the leakage of pre-mRNAs.
Discussion
The mechanisms of nuclear retention of pre-mRNAs have been extensively studied in yeast. A pioneer study by Legrain and Rosbash (25) revealed that early intron recognition is required for the nuclear retention of pre-mRNAs. Accordingly, mutations in the 5′ splice site and branchpoint and mutations in the early splicing factors lead to the leakage of pre-mRNA to the cytoplasm (25–27). Thus formation of early splicing complexes appears to be responsible for the nuclear retention, and it has therefore been difficult to separate nuclear retention and splicing. Rutz and Seraphin (28) showed that particular mutations in the yeast branch point binding protein (BBP/SF1) gene induce leakage of pre-mRNAs to the cytoplasm without affecting the splicing activity in vitro. More recently, Galy et al. (29) showed that depletion of MLP1, a constituent of the fiber-like structures emanating from the nuclear pore complexes, impairs retention of pre-mRNAs without having an effect on splicing in vivo. This was the first demonstration of clear separation of the two processes. However, how MLP1 is related to the early intron recognition is not well understood. A trimeric protein complex, termed RES (pre-mRNA retention and splicing), was also implicated in pre-mRNA nuclear retention and splicing in yeast (30).
The mechanisms of nuclear retention of pre-mRNAs have been much less studied and are less well understood in vertebrates than in yeast, although some earlier work showed that early intron recognition is responsible (31). Importantly, some viruses such as HIV-1, HTLV-1, and the type D retroviruses have systems to induce nuclear export of intron-containing viral pre-mRNAs, and these phenomena have been extensively studied (reviewed in ref. 8). However, at least in terms of the nuclear retention mechanisms of cellular pre-mRNAs, these studies have not so far given much more insight than the studies in yeast.
One feature of vertebrate splicing is ambiguity of the splicing signals. Because splice site sequences are less conserved, vertebrates often use additional signals for splicing that are missing in yeast (reviewed in ref. 32). One such signal is the purine-rich ESE that stimulates splicing of the adjacent upstream intron (reviewed in refs. 11–13). ESEs are involved in the regulation of splicing and are frequently found in alternatively spliced exons (33, 34). Our results showed that ESEs per se possess an activity to retain RNAs in the nucleus. Moreover, this retention activity of ESEs shares a saturatable nuclear factor with the nuclear retention of regular pre-mRNAs. Thus, we have obtained a clue to separate nuclear retention from intronic sequences. ESEs should be a useful tool for studying nuclear retention of pre-mRNAs. We also showed that the splicing reaction appeared to release or reset the nuclear retention state caused by ESEs, leading to nuclear export of mature mRNAs. How then are nuclear retention and export coordinated?
Fig. 5 shows our current model of the processes. ESE sequences are bound by SR protein family (15–17) but probably as a part of a larger complex containing other factors. This complex retains the pre-mRNA and stimulates splicing concomitantly. At least a part of splicing stimulation should be achieved by the addition of U2 snRNP onto the branchpoint in the upstream intron (11, 12). Consistently, we have recently shown that 17S U2 snRNP is associated with ESEs in Xenopus oocyte nucleus (35). It is unlikely, however, that 17S U2 snRNP per se is the nuclear retention factor because depletion of U2 did not induce export of ESE-containing RNAs or pre-mRNAs, although it is plausible that U2 contributes partly to the nuclear retention. It should be pointed out that our earlier work showed that depletion of even both U1 and U2 did not induce export of pre-ftz (9). Concomitantly with splicing, the exon junction complex (EJC) and the transcription/export complex (TREX) are imprinted onto the RNA (Fig. 5 and refs. 36 and 37). The retention factor bound to ESEs is either transferred onto the excised intron or inactivated, thereby releasing the retention of the spliced mRNA. TAP/p15 is recruited to the spliced mRNA through the EJC and/or TREX, leading to efficient export (36, 38, 39), whereas the excised intron is retained in the nucleus and eventually degraded.
Fig. 5.
Model of action of ESEs. How ESEs may contribute to gene expression and RNA quality control is illustrated. See Discussion for details.
What is the identity of the retention factors? Several factors, including SR proteins, U2AF, and components of SF3a/b complexes, were shown to be directly or indirectly associated with ESEs (15–17, 35). It is possible that some of these factors exert the nuclear retention activity, independently of U2 snRNP per se. For instance, ESE-bound SR proteins, especially their phosphorylated forms, may be responsible. It has been proposed that phosphorylated SR proteins on the RNA should be either dephosphorylated or exchanged with other dephosphorylated shuttling SR proteins, concomitantly with splicing reaction, to be able to induce RNA export (23, 40, 41). Dephosphorylation of SR proteins may be the switch to release the nuclear retention. However, our preliminary results suggest that SR proteins per se are not the saturable retention factors because the amount of the GAA competitor used in our experiments was not sufficient to saturate highly abundant SR proteins (data not shown). Alternatively, early splicing factors that are implicated in the nuclear retention in yeast (such as BBP/SF1) are also good candidates. There is no evidence, however, that these early splicing factors are tightly associated with ESEs. Further experiments are required to elucidate the molecular mechanism of nuclear retention of ESEs and pre-mRNAs.
Materials and Methods
DNA Constructs.
For U1 derivative plasmids, annealed oligo DNA fragments designed to produce XhoI and SalI sites were inserted into the XhoI site of T7 Xenopus U1ΔSm plasmid. For β-globin derivative plasmids, ESE fragments were amplified by PCR from U1-derivative plasmids and then inserted into the BamHI and EcoRI sites of the plasmid containing the β-globin cDNA sequence or the β-globin sequence with an intron (10).
In Vitro Transcription.
32P-labeled RNAs were transcribed as described (10) except that only [α-32P]UTP was used. The transcription reaction was usually performed at 37°C for 45–60 min. Unlabeled RNAs were synthesized by using MEGAscript (Ambion, Austin, TX) according to the manufacturer's instructions.
RNA Microinjection into Xenopus Oocytes.
RNA microinjection into Xenopus oocytes was performed as described (10). For preparation of BSA-NES and BSA-mut, PKI NES peptide (CELALKLAGLDIN) or a mutant peptide (CELALKAAGADIN) was conjugated to BSA as described (10) Analysis and quantitation of RNA bands were performed with BAS-2500 (Fuji Film, Tokyo, Japan) and Image Gauge version 3.45 (Fuji Film).
Northern Blot Analysis.
RNA samples were fractionated by denaturing PAGE and transferred to hybond N+ membrane (Amersham, Piscataway, NJ). Hybridization with oligo DNA probes,32P-labeled at the 5′ termini, was performed in PerfectHybPlus (Sigma, St. Louis, MO). Oligo DNAs TGCAGTCGAGTTTCCCGCA and AGATACTACACTTGATCTTAGC were used as the probes for U1 and U2 snRNAs, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Akiya Watakabe (National Institute for Basic Biology, Okazaki, Aichi, Japan) for ESE plasmids, Dr. Iain Mattaj for helpful comments on the manuscript, and members of our laboratory for suggestions and criticisms of this manuscript. This work was supported by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency and grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. I.T. and K.M. were supported by the 21st Century Centers of Excellence Program of the Ministry of Education, Culture, Sports, Science, and Technology of Japan. K.M. was a Japan Society for the Promotion of Science Research Fellow.
Abbreviations
- ESE
exonic splicing enhancer
- snRNA
small nuclear RNA
- ASLV
avian sarcoma-leukosis virus
- cTNT
cardiac troponin T
- β-TM
β-tropmyosin
- DHFR
dihydrofolate reductase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.I. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704922104/DC1.
References
- 1.Tran EJ, Wente SR. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Vinciguerra P, Stutz F. Curr Opin Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 3.DeCerbo J, Carmichael GG. Curr Opin Cell Biol. 2005;17:302–308. doi: 10.1016/j.ceb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Wolin SL, Cedervall T. Annu Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 5.Henras AK, Dez C, Henry Y. Curr Opin Struct Biol. 2004;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Saguez C, Olesen JR, Jensen TH. Curr Opin Cell Biol. 2005;17:287–293. doi: 10.1016/j.ceb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Sommer P, Nehrbass U. Curr Opin Cell Biol. 2005;17:294–301. doi: 10.1016/j.ceb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Cullen BR. Trends Biochem Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 9.Ohno M, Segref A, Kuersten S, Mattaj IW. Mol Cell. 2002;9:659–671. doi: 10.1016/s1097-2765(02)00454-9. [DOI] [PubMed] [Google Scholar]
- 10.Masuyama K, Taniguchi I, Kataoka N, Ohno M. Genes Dev. 2004;18:2074–2085. doi: 10.1101/gad.1216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blencowe BJ. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 12.Reed R. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 13.Cartegni L, Chew SL, Krainer AR. Nat Rev. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka K, Watakabe A, Shimura Y. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tacke R, Tohyama M, Ogawa S, Manley JL. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 16.Sun Q, Mayeda A, Hampson RK, Krainer AR, Rottman FM. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 17.Kondo S, Yamamoto N, Murakami T, Okumura M, Mayeda A, Imaizumi K. Genes Cells. 2004;9:121–130. doi: 10.1111/j.1356-9597.2004.00709.x. [DOI] [PubMed] [Google Scholar]
- 18.Shen H, Green MR. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Lam BJ, Hertel KJ. RNA. 2002;8:1233–1241. doi: 10.1017/s1355838202028030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robberson BL, Cote GJ, Berget SM. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Gattoni R, Stevenin J, Steitz JA. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Steitz JA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Yario TA, Steitz JA. Proc Natl Acad Sci USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai MC, Tarn WY. J Biol Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- 25.Legrain P, Rosbash M. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 26.Rain JC, Legrain P. EMBO J. 1997;16:1759–1771. doi: 10.1093/emboj/16.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luukkonen BG, Seraphin B. Nucleic Acids Res. 1999;27:3455–3465. doi: 10.1093/nar/27.17.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutz B, Seraphin B. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 30.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang DD, Sharp PA. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 32.Smith CW, Valcarcel J. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 33.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 34.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Masuyama K, Taniguchi I, Okawa K, Ohno M. Biochem Biophys Res Commun. 2007;359:580–585. doi: 10.1016/j.bbrc.2007.05.144. [DOI] [PubMed] [Google Scholar]
- 36.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 37.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 40.Lin S, Xiao R, Sun P, Xu X, Fu XD. Mol Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Gilbert W, Guthrie C. Mol Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.