Abstract
We have identified a conserved region in the C-terminal domain of bromodomain-containing protein 4 (BRD4) that mediates its specific interaction with positive transcription elongation factor b (P-TEFb). This domain is highly conserved in testis-specific bromodomain protein (BRDT) and Drosophila fs(1)h. Both BRDT and fs(1)h specifically interact with P-TEFb in mammalian cells, and this interaction depends on their C-terminal domains. Overexpression of the BRD4 P-TEFb-interacting domain disrupts the interaction between the HIV transactivator Tat and P-TEFb and suppresses the ability of Tat to transactivate the HIV promoter. Incubation of cells with a synthetic peptide containing the C-terminal domain of BRD4 interferes with transactivation of the HIV promoter by the Tat protein.
Keywords: cyclin-dependent kinase 9
Although great progress has been made in eliminating productive HIV infection in patients, a small fraction of resting CD4+ memory T cells harbor transcriptionally silent HIV provirus that can serve as a source of reemergent virus after cessation of highly active antiretroviral therapy. Given the exceptionally long life span of memory T cells, it appears unlikely that the virus will be eliminated by conventional antiretroviral therapy alone (1).
The mechanisms leading to transcriptional silencing and proviral latency are incompletely understood. The majority of productive and latent integrations appear to occur within the coding regions of active genes (2, 3). During a recent study examining the site of HIV integration in latently infected cells, we identified a number of “hot-spot” genes that were the target of multiple independent integrations in latent but not productively infected populations (3). It is unlikely that these latent integrations clustered by chance, suggesting the possibility that these genes may somehow be involved in HIV transcription and that perhaps retroviral integration disrupts their function, leading to HIV latency.
By comparison of productive and latent integration sites, we identified the bromodomain-containing protein 4 (BRD4) gene as a hot spot for HIV integration specifically in latent populations. Four independent integrations were identified within the first intron of the BRD4 gene. BRD4 is a member of the BET family of proteins, which characteristically have two tandem N-terminal bromodomains followed by an extraterminal (ET) domain (4). The bromodomain is an acetyllysine-binding motif, and BRD4 has been shown to associate with acetylated histones (5). BRD4 has also been identified as an interaction partner with the positive transcription elongation factor b (P-TEFb) complex, a factor crucial for efficient HIV transcriptional elongation and expression (6–8). P-TEFb is a heterodimer of cyclin-dependent kinase 9 (CDK9) and cyclin T1, T2, or K. Recruitment of P-TEFb to the HIV promoter by the viral transactivator Tat leads to phosphorylation of the C-terminal domain (CTD) of RNA polymerase II and more robust transcriptional elongation (6).
In the following set of experiments, we determine the region of interaction between BRD4 and P-TEFb and its effect on HIV transcription, and we describe a peptide that transduces into cells and inhibits HIV transcription.
Results
BRD4 Interacts with P-TEFb Through a Short CTD.
To identify the portion of BRD4 that interacts with P-TEFb, we performed coimmunoprecipitation experiments with FLAG-tagged BRD4 deletion mutants. 293T cells were transfected with expression constructs encoding either the short isoform of BRD4(1–722), the long isoform (1–1362), mutants lacking both bromodomains (1362ΔBD1&2), the ET domain (1362ΔET), or the C-terminal half of the protein (722–1362). After anti-FLAG immunoprecipitation, BRD4-associated P-TEFb was detected by immunoblotting with antisera specific for cyclin T1 or CDK9. P-TEFb was not detected in immunoprecipitates of the N-terminal half of BRD4(1–722), suggesting that the association occurs in the C-terminal half of BRD4 (Fig. 1A). Deletion of the bromodomains in the full-length BRD4 (1362ΔBD1&2) had little effect on P-TEFb interaction, suggesting that their association is not mediated by these domains. Deletion of the ET domain appeared to decrease modestly the level of interaction between P-TEFb and BRD4; however, this deletion mutant was also expressed as lower levels (Fig. 1A). The ET domain is a poorly defined domain thought to mediate protein–protein interactions (4). Interestingly, the C-terminal half of BRD4(722–1362) is sufficient to immunoprecipitate P-TEFb. This region of BRD4 is missing in the short isoform and contains no known functional motifs (Fig. 1A).
Fig. 1.
BRD4 interacts with P-TEFb by means of a short CTD. (A) FLAG-tagged deletion mutants of BRD4 were overexpressed in 293T cells. Nuclear extracts (NE) and anti-FLAG immunoprecipitates (IP) were immunoblotted (IB) for CDK9 and cyclin T1. The small arrows point to the full-length proteins, and the smaller bands likely reflect degradation products. (B) FLAG-tagged BRD4 constructs encoding residues 1209–1362, 1209–1362Δ1279–1304, 1209–1362Δ1306–1330, 1209–1362Δ1329–1362, 1209–1362Δ1329–1345, and 1209–1362Δ1345–1362 were overexpressed in 293T cells. Anti-FLAG immunoprecipitates were immunoblotted with anti-CDK9 or anti-cyclin T1.
The region of P-TEFb interaction was narrowed to the C-terminal 153 residues of BRD4 by further deletion experiments (data not shown). Scanning deletion analysis was performed to pinpoint the interacting region. Expression constructs encoding FLAG-tagged BRD4 mutants were introduced into 293T cells by calcium phosphate transfection, and coimmunoprecipitation of CDK9 was detected by Western blotting. Deletion of amino acids 1279–1304 had a modest effect on P-TEFb interaction, although this construct was also expressed at a lower level than the wild-type construct (1209–1362) (Fig. 1 A and B). Deletion of amino acids 1306–1330 of BRD4 had an even more deleterious effect on P-TEFb interaction with barely detectable levels of CDK9 coimmunoprecipitation. Deletion of the 1329–1362, 1329–1345, and 1345–1362 regions completely eliminated the interaction of P-TEFb with BRD4. Thus, the C-terminal 34 aa of BRD4 are crucial for interaction with P-TEFb.
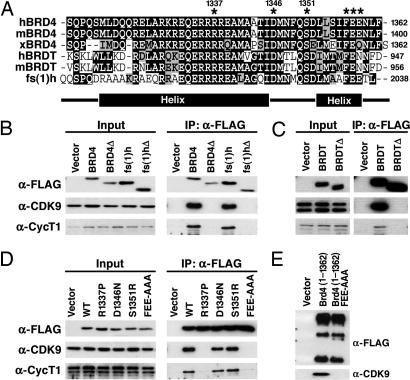
Analysis of the sequence of the CTD of BRD4 against protein databases revealed the conservation of this domain with two other members of the BET family: testis-specific bromodomain protein (BRDT) and Drosophila female sterile homeotic [fs(1)h]. This conservation was recently noted independently (9). Fig. 2A shows an multialignment of the CTD of human, mouse, and Xenopus BRD4; human and mouse BRDT; and Drosophila melanogaster fs(1)h. Compared with human and mouse BRD4, this region in Xenopus BRD4 is 90% conserved, human and mouse BRDT are both 75% conserved, and Drosophila fs(1)h is 69% conserved. To determine whether the CTD of fs(1)h and BRDT also interact with P-TEFb, FLAG-tagged expression constructs for human BRD4, Drosophila fs(1)h, and murine BRDT as well as C-terminal deletion mutants of each, were introduced into 293T cells by transient transfection. Nuclear [BRD4 and fs(1)h] or whole-cell (BRDT) extracts were prepared, and α-FLAG immunoprecipitations were carried out. Coimmunoprecipitation of endogenous (human) P-TEFb revealed that CDK9 and cyclin T1 associated with BRD4, fs(1)h, and BRDT but not C-terminal deletion mutants [BRD4Δ, fs(1)hΔ, and BRDTΔ, respectively] lacking the equivalent P-TEFb interacting region of BRD4 (Fig. 2 B and C). This result points to the functional importance of the CTD of these proteins as a mediator of P-TEFb interaction and also suggests that its structure is highly conserved within select members of the BET family.
Fig. 2.
A C-terminal peptide conserved among BRD4, BRDT, and fs(1)h mediates P-TEFb interaction. (A) Alignment of the C termini of human, murine, and Xenopus BRD4; human and murine BRDT; and Drosophila fs(1)h. Identical residues are indicated in black, and conserved residues are in gray. The locations of α-helices predicted by computer analysis are indicated below. (B) Vectors encoding the C termini of human BRD4(1209–1362), BRD4(Δ1209–1344), fs(1)h(1885–2038), or fs(1)hΔ(1885–2020) were transfected into 293T cells. Anti-FLAG immunoprecipitates (IP) were immunoblotted with anti-FLAG, anti-cyclin T1 (anti-CycT1), and anti-CDK9. (C) Expression constructs encoding the C terminus of BRDT (residues 821–956) or a C-terminal deletion mutant [BRDTΔ(821–938)] were transfected into 293T cells. Whole-cell lysates were prepared, and BRDT was immunoprecipitated with anti-FLAG beads. (D) C-terminal residues conserved among BRD4, BRDT, and fs(1)h were mutated in FLAG-BRD4(1209–1362) (indicated by an asterisk in A). Plasmids encoding BRD4 point mutants 1209–1362R1337P, D1346N, S1351R, and FEE1357AAA were transfected into 293T cells. Anti-FLAG immunoprecipitates were immunoblotted for FLAG, CycT1, and CDK9. (D) Plasmids encoding FLAG tagged wild-type (WT) BRD4 (1–1362) or mutant (1–1362FEE) were transfected into 293T cells. Anti-FLAG immunoprecipitates were blotted for anti-FLAG and anti-CDK9.
To isolate mutants of BRD4 that do not interact with P-TEFb, we prepared a series of point mutants in the C terminus of human BRD4(1209–1362) based on residues conserved among BRD4, BRDT, and fs(1)h (Fig. 2A). By coimmunoprecipitation, the single-point mutants D1346N and S1351R had no effect on the interaction of BRD4 and P-TEFb (Fig. 2D). However, replacing the arginine at position 1337 with proline (R1337P) disrupted the BRD4–P-TEFb interaction. Similarly, conversion Phe and Glu to Ala (F1357A/E1358A/E1359A) eliminated its interaction with P-TEFb. Introduction of the FEE mutation into full-length BRD4 eliminated any interaction with P-TEFb, indicating that the C terminus of BRD4 is essential for this interaction (Fig. 2E).
BRD4 Interacts with Both Subunits of P-TEFb.
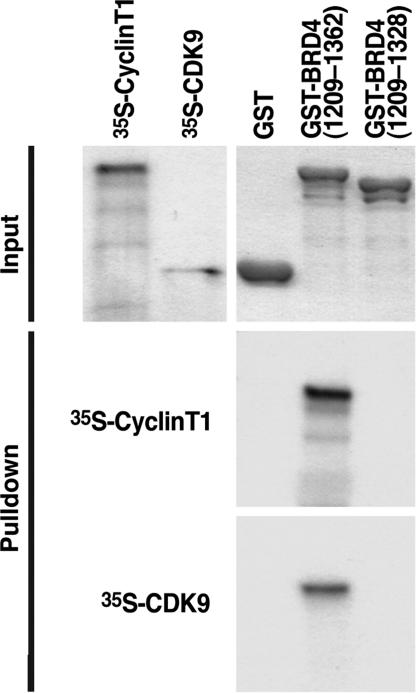
To determine which subunit of P-TEFb mediates the interaction with BRD4, we performed GST pulldown experiments. GST fusion proteins representing either GST alone, GST-BRD4(1209–1362), or the deletion mutant GST-BRD4(1209–1328) were overexpressed and purified from bacterial lysates. Washed beads coated with each GST fusion protein were incubated with either in vitro translated 35S-radiolabeled human cyclin T1 or CDK9. After extensive washing, interacting proteins were eluted, visualized by Coomassie blue staining after SDS/PAGE, and processed for autoradiography. Both in vitro translated cyclin T1 and CDK9 interacted with GST-BRD4(1209–1362) but not with GST-BRD4(1209–1328) (Fig. 3). These data indicate that each subunit of P-TEFb directly and independently interacts with BRD4.
Fig. 3.
BRD4 interacts with both P-TEFb subunits, CDK9, and cyclin T1. GST pulldowns were performed by incubating GST-, GST-BRD4(1209–1362), or GST-BRD4(1209–1328)-coated agarose beads with 35S-radiolabeled human cyclin T1 or human CDK9. After extensive washing, proteins were eluted with Laemmli buffer, separated by SDS/PAGE, and visualized by Coomassie blue staining and autoradiography.
Direct Recruitment of P-TEFb to the GAL4 Promoter by BRD4.
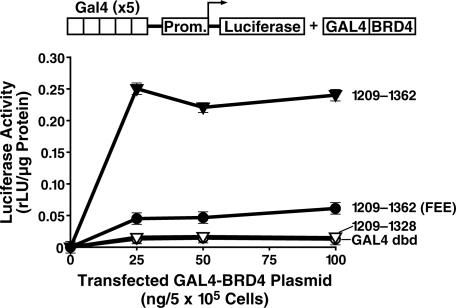
These results indicate that the C terminus of BRD4 interacts with P-TEFb. To determine its possible role as a transactivation domain, we tethered the C terminus of BRD4 to the DNA-binding domain of GAL4. Although expression of the GAL4 DNA-binding domain alone had no effect on a 5×GAL4-UAS-luciferase reporter construct (Fig. 4), overexpression of the wild-type GAL4-BRD4 fusion (BRD4 amino acids 1209–1362) led to robust transcriptional activation (Fig. 4). The FEE to AAA point mutant and deletion mutant (1209–1328) had minimal transactivation activity. These results indicate that the CTD of BRD4 functions as an autonomous transactivation domain.
Fig. 4.
The P-TEFb-interacting domain (PID) of BRD4 is an autonomous transactivation domain. (A) Increasing amounts of plasmids encoding the GAL4 DNA-binding domain (dbd) fused to BRD4(1209–1362), BRD4(1209–1328), or BRD4(1209–1362)(FEE1357AAA), were cotransfected with p5×GAL4-UAS-luciferase into HeLa cells. After 36 h, lysates were prepared, and luciferase levels were determined.
BRD4 Overexpression Inhibits Tat-Mediated HIV Transactivation.
The viral transactivator Tat plays a critical role in HIV transcription because of its ability to recruit the P-TEFb complex to the HIV promoter. Recruitment of P-TEFb by Tat markedly enhances the processivity of the HIV promoter-associated RNA polymerase II complex. To determine the effect of BRD4 expression of HIV transcription, we tested its effect on a HIV transcriptional reporter in the presence of Tat. Overexpression of full-length BRD4 inhibited Tat-mediated HIV transcription, suggesting that it competes with Tat for P-TEFb (Fig. 5B). Similarly, overexpression of the C-terminal 153 aa (1209–1362) or 102 aa (1260–1362) of BRD4 interfered with HIV transcription to an even greater extent than the full-length isoform.
Fig. 5.
Inhibition of Tat transactivation of the HIV LTR by the PID of BRD4. (A) Diagram showing the constructs used in this experiment: full-length BRD4, BRD4(1209–1362), and BRD4(1260–1362). (B) HeLa cells were transfected with 25 ng of HIV LTR-luciferase reporter and 5 ng of Tat expression plasmid and increasing amount of BRD4 expression plasmids (0–400 ng).
BRD4 Suppresses the Tat–P-TEFb Interaction.
During Tat transactivation, cyclin T1 is recruited to the HIV LTR through Tat and the TAR element found at the 5′ end of all viral transcripts. This association brings CDK9 to the LTR and promotes transcriptional elongation by enhancing phosphorylation of the CTD of RNA polymerase II. To determine the mechanism whereby BRD4 interferes with this process, we performed in vitro competition experiments to assess the effect of BRD4 on Tat–P-TEFb interaction (Fig. 6). Binding reactions consisting of a constant amount of biotinylated synthetic Tat and recombinant P-TEFb as well as varying amounts of in vitro translated BRD4(1209–1362) or BRD4(1209–1328) were prepared. The ability of Tat to bind to P-TEFb (CDK9) in the presence of BRD4 peptides was measured by Western blotting (α-CDK9) after binding of Tat to streptavidin/agarose and centrifugation. Increasing amounts of BRD4(1209–1362) but not BRD4(1209–1328) decreased the amount of P-TEFb (CDK9) that interacted with Tat (Fig. 6). This result indicates that BRD4 expression interferes with Tat binding to P-TEFb and suggests that BRD4 could inhibit the Tat-mediated recruitment of P-TEFb to the viral LTR.
Fig. 6.
The PID of BRD4 inhibits the binding of Tat to P-TEFb. Biotinylated Tat was incubated with recombinant P-TEFb in the presence of BRD4 peptides corresponding to either amino acids 1209–1362 or 1209–1328 obtained after in vitro transcription/translation. Biotinylated Tat and associated factors were pulled down by streptavidin/agarose followed by Western blotting with an antiserum specific for CDK9 or streptavidin/horseradish peroxidase (SA-HRP) (to verify equal pulldown of Tat). The BRD4 peptides are visualized by autoradiography after SDS/PAGE.
Exogenous BRD4 Peptide Interferes with Tat Transactivation.
To test this possibility, we synthesized a peptide that fuses the protein transduction domain (PTD) of Tat to the C terminus of BRD4 (Fig. 7A). The PTD confers receptor- and energy-independent entry of fused proteins into cells. Increasing amounts of either the PTD peptide alone, the PTD-BRD4 peptide, or the chemical CDK9 inhibitor, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) were added to HeLa cells transfected with an HIV LTR-luciferase reporter and a Tat expression plasmid. HIV transcription was inhibited 6-fold by increasing amounts of PTD-BRD4 peptide but not the PTD peptide alone. As expected, similar results were obtained with the DRB control.
Fig. 7.
A BRD4 PID peptide suppresses Tat transactivation of the HIV LTR and reactivation from latency. (A) A synthetic peptide called PTD-BRD4 consists of the protein transduction domain of Tat (Tat-PTD) followed by BRD4(1318–1362). (B) HeLa cells were transfected with 25 ng of HIV LTR-luciferase with (+) or without (−) 5 ng of Tat expression plasmid. The PTD alone (pale gray), PTD-BRD4 peptide (gray), or DRB (black) was added to the cell-culture supernatant at concentrations ranging from 0.01 to 10 μM. Luciferase activities were assayed 24 h after transfection. (C) Clonal cell line A2 contains a single latent HIV provirus and expresses GFP after reactivation of HIV by TNF-α treatment. A2 cells were pretreated for 2 h with either the PTD-BRD4 peptide (gray circles), PTD alone (white circles), or DRB (black triangles) at the indicated concentrations, followed by the addition of TNFα (10 ng/ml). After 18 h, levels of HIV proviral transcription were measured by FACS for GFP.
To confirm this result, we tested the ability of the PTD-BRD4 peptide to interfere with the reactivation of HIV transcription in a latently infected cell line, the J-Lat clone A2 (10). Increasing amounts of PTD-BRD4 peptide or DRB, but not the PTD alone, interfered with the TNF-α-induced reactivation and GFP expression in this clonal cell line (Fig. 7C).
Discussion
We have identified a conserved domain in the CTD of BRD4, BRDT, and Drosophila fs(1)h that mediates the selective recruitment of the P-TEFb complex. We propose to call this domain the PID. This interaction is highly conserved because both Drosophila fs(1)h and murine BRDT interacted strongly with human P-TEFb in coimmunoprecipitation experiments. Overexpression of the PID disrupts the interaction between Tat and P-TEFb and interferes with Tat-transactivating activity on the HIV promoter. Furthermore, we have characterized a synthetic peptide corresponding to the PID capable of directly transducing cells and interfering with Tat-induced transactivation of the HIV promoter.
It has been proposed that BRD4 may function in “transcriptional memory” to maintain proper gene expression patterns during cell division. This hypothesis stems from the finding that BRD4 binds to acetylated (generally active) chromatin through its bromodomains and remains chromatin-bound during mitosis (5). BRD4 could therefore be targeted to regions of active (acetylated) chromatin through its bromodomains, where it could activate transcription by PID-mediated P-TEFb recruitment after cell division. Our data support this notion because we have shown that the PID can activate transcription when tethered to a heterologous promoter. This finding suggests that BRD4 may function as a molecular adapter with its bromodomains involved in its targeting to acetylated chromatin, whereas the PID serves to recruit P-TEFb.
Our findings differ significantly from previously published results, indicating that the bromodomains of murine BRD4 are crucial for its interaction with P-TEFb (8). We found little difference in the level of interaction of BRD4 with P-TEFb between wild-type BRD4 and a BRD4 construct lacking both bromodomains (8). A possible reason for these differing observations could be that we used a construct that precisely deleted the two bromodomains, whereas additional neighboring sequences were deleted in the previously published constructs. It is possible that these larger deletions resulted in protein misfolding or mislocalization, preventing their interaction with P-TEFb. The same group also reported that the N-terminal half of BRD4 was sufficient to interact with P-TEFb. However, sequence analysis of this construct (f-ΔC-term) indicates that it actually contains an internal deletion of residues 700-1317 of BRD4, resulting in an in-frame fusion of the N-terminal half (1–699) to the CTD (1318–1400) of BRD4 (data not shown). This fusion protein therefore contains the PID that we identify here and further supports our observations.
BRD4 appears to be intimately involved in the life cycle of human papillomavirus (HPV). BRD4 specifically interacts with the HPV E2 protein and thereby tethers the viral genome to host chromatin, enabling its partition in daughter cells during mitosis (11–13). Interestingly, the region of BRD4 that interacts with E2 has recently been shown to be the PID (14). Because both E2 and P-TEFb interact with the same C-terminal domain of BRD4, it will be important in future experiments to determine whether both proteins can bind to BRD4 at the same time or whether they compete for binding to BRD4. Because E2 is a transcriptional regulator, competition for the PID will likely have important implications for regulation of HPV transcription. The crystal structure of the C-terminal 20 residues of BRD4 bound to E2 shows the C-terminal domain of BRD4 to be a highly α-helical structure. The FEE to AAA mutation that we report here and that abrogates P-TEFb interaction is located in the C-terminal end of the helix. The effect of the R1337P mutation is less clear because this region of BRD4 was not part of the solved structure. However, computer analysis predicts that this amino acid lies in the middle of a second upstream helical region, and substitution of a proline in this helix is predicted to disrupt its helical nature.
Beyond the conservation of the PID among BRD4, BRDT, and fs(1)h, it is intriguing to note that each of these genes encodes two alternative splice products encoding the N-terminal half of the protein (containing the bromo and ET domains) but lacking the C-terminal half containing the PID. This shorter isoform, lacking the PID, could potentially function as a dominant-negative isoform capable of interfering with the recruitment of P-TEFb to acetylated chromatin by competing with the longer isoform. It will be interesting to determine whether the alternative splicing is differentially regulated and how the levels of each isoform vary. Another potential function for the short isoform is suggested from experiments with the short isoform of BRDT, indicating that it plays a role in chromatin compaction during spermatogenesis (15).
Little is known about the molecular function of fs(1)h in Drosophila. Genetic screens initially identified fs(1)h as a maternal effect gene and suggested a role in segment identity determination (16). Loss of fs(1)h function during embryogenesis altered the expression of several genes including Kruppel (Kr), eve, en, and Ultrabithorax (Ubx), leading to homeotic transformations (17). Because our data suggest that the C-terminal domain of fs(1)h also recruits P-TEFb, changes in gene expression in fs(1)h mutants might be caused by mislocalization of P-TEFb and subsequent failure to activate appropriate transcription. It will be interesting to determine whether point mutations in the PID of fs(1)h interfere with its ability to complement fs(1)h mutant embryos. Failure to restore proper gene expression and correct the segmentation defects observed in fs(1)h mutants would suggest that recruitment of P-TEFb by fs(1)h is an important step during embryogenesis.
We have shown that a BRD4 PID peptide inhibits HIV transcription by interfering with the Tat-mediated recruitment of P-TEFb, a step necessary for high-level viral gene expression. As discussed above, we have identified four independent HIV integration events in the BRD4 gene in latently infected cell lines (3). We therefore hypothesize that integration of HIV in the BRD4 gene led to an increase in BRD4 expression, resulting in the suppression of HIV expression. Although we do not have individual clonal cell lines to test this hypothesis directly, future experiments will examine in greater detail the potential role of BRD4 in HIV latency.
The PID is an especially interesting therapeutic target because it appears to play a role in the life cycles of two important human pathogens, HIV and HPV. Abbate et al. (14) recently reported that a BRD4 peptide similar to ours disrupts the E2–BRD4 interaction. As discussed above, it will be important to determine whether E2 and P-TEFb can bind simultaneously to BRD4 or whether E2 competes with P-TEFb for binding to BRD4. Our observations also warrant a careful evaluation of the role of P-TEFb in the biology of HPV replication.
Experimental Procedures
Cell Lines and Plasmids.
Jurkat, HeLa, and 293T cells were obtained from the American Type Culture Collection (Manassas, VA). ORFs encoding the short (amino acids 1–722) and long (amino acids 1–1362) isoforms of human BRD4 were isolated by RT-PCR from leukocyte cDNA (Clontech, Palo Alto, CA) and Jurkat cDNA, respectively, and inserted into pFLAG-CMV2 (Sigma–Aldrich, St. Louis, MO). Deletion mutants were cloned by PCR (oligonucleotide sequences available upon request). Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Drosophila fs(1)h and murine BRDT were cloned by RT-PCR from whole Drosophila cDNA and mouse testes cDNA, respectively.
Immunoprecipitation.
FLAG-tagged BRD4, fs(1)h, and BRDT constructs were introduced into 293T cells by calcium phosphate-mediated transfection. Nuclear extracts [BRD4, fs(1)h)] were prepared by hypotonic lysis and high-salt extraction (18). For BRDT, whole-cell lysates were prepared by direct lysis in a modified buffer C [20 mM Hepes, pH 7.9/1.5 mM MgCl2/25% (vol/vol) glycerol/420 mM NaCl/0.2 mM EDTA/0.2% Nonidet P-40/0.5 mM DTT]. Lysates were dialyzed into dialysis buffer [20 mM Hepes/20% (vol/vol) glycerol/150 mM NaCl/0.2 mM EDTA/0.5 mM DTT/1 mM PMSF]. FLAG-tagged proteins were immunoprecipitated with anti-FLAG-agarose beads (Sigma–Aldrich). Coimmunoprecipitating proteins were detected by Western blotting with rabbit anti-FLAG (Sigma–Aldrich), anti-cyclin T1, or anti-CDK9 (Santa Cruz Biotechnology, Santa Cruz, CA).
GST Pulldowns.
GST, GST-BRD4(1209–1362), and GST-BRD4(1209–1328) were overexpressed in BL21 and purified on GST-Sepharose beads as recommended by the manufacturer (GE Healthcare, Piscataway, NJ). Human cyclin T1- and CDK9-encoding RNAs were prepared with the T7 Ribomax Express in vitro transcription kit (Promega, Madison, WI). Radiolabeled cyclin T1 and CDK9 were prepared by in vitro translation with [35S]methionine and RNase-treated rabbit reticulocyte lysates. GST pulldowns were performed by incubating washed beads coated with either GST, GST-BRD4(1209–1362), and GST-BRD4(1209–1328) with radiolabeled cyclin T1 or CDK9. After extensive washing, purified proteins were detected by Coomassie blue staining and fluorography (Amplify; GE Healthcare).
Inhibition of Tat–P-TEFb Interaction by BRD4.
Synthetic biotinylated tat was incubated with purified human P-TEFb (Millipore, Bedford, MA), in vitro translated, 35S-labeled BRD4(1209–1362), or BRD4(1209–1328), and streptavidin-conjugated agarose beads. In vitro translated BRD4 was diluted 5- or 25-fold with rabbit reticulocyte lysate where indicated. Unbound proteins were removed by extensive washing [20 mM Hepes/20% (vol/vol) glycerol/150 mM NaCl/0.2 mM EDTA/0.5 mM DTT/0.1% Nonidet P-40]. Input levels of BRD4 were detected by fluorography with Amplify. CDK9 and biotinylated Tat were detected by Western blotting with either anti-CDK9 or streptavidin/horseradish peroxidase.
Luciferase Assays.
For the GAL4 transactivation experiments, HeLa cells were cotransfected with p5×GAL4-UAS-luciferase reporter plasmid and increasing amounts of GAL4-BRD4(1209–1362), 1209–1362FEE, or 1209–1328. Cell lysates were prepared and luciferase activity analyzed by using the dual-luciferase reporter assay system (Promega). Inhibition of HIV transcription was measured by transfecting HeLa cells with 25 ng of HIV LTR-luciferase reporter, 5 ng of pRSV-Tat, and 0–400 ng of pFLAG-CMV2-BRD4 expression plasmids (FuGENE 6; Roche, Indianapolis, IN). The total amount of DNA was kept constant with empty pFLAG-CMV2.
Inhibition of HIV transcription with Tat PTD-BRD4 peptide was shown by transfecting HeLa cells with 25 ng of HIV LTR-luciferase reporter, 5 ng of pRSV-Tat, and 400 ng of pFLAG-CMV2 per well. Chemically synthesized HPLC-purified peptides (Tat PTD-BRD4 or Tat-PTD alone) were then added to the cell culture supernatant at a final concentration of 0, 0.1, 1, or 10 μM. Control reactions with the P-TEFb inhibitor DRB were performed in parallel.
Inhibition of Latent HIV Transactivation.
Latent HIV clone A2 contains a single latent HIV provirus that expresses Tat and GFP upon reactivation (10). A2 cells were preincubated for 2 h with 2–10 μM Tat PTD-BRD4(1209–1362), Tat PTD alone, or DRB. TNF-α was then added to the cells at 10 ng/ml, and reactivation of the latent HIV provirus was detected 18 h later by FACS analysis (FACSCalibur; BD Bioscience, San Jose, CA).
Acknowledgments
We thank Amanda Bradford for manuscript preparation, John Carroll for graphics, and Steve Ordway and Gary Howard for editorial assistance. We thank Rick Bushman (University of Pennsylvania, Philadelphia, PA) and Mary Lewinski (The Salk Institute, La Jolla, CA) for sharing the sequence of the latent HIV integration sites. This research was supported by the National Institutes of Health (to E.V.) and the University of California, San Francisco-Gladstone Institute of Virology and Immunology Center for AIDS Research Grant P30 AI27763 (to D.A.B.).
Abbreviations
- BRD4
bromodomain-containing protein 4
- BRDT
testis-specific bromodomain protein
- CDK9
cyclin-dependent kinase 9
- CTD
C-terminal domain
- DRB
5,6-dichloro-1-β-l-ribofuranosylbenzimidazole
- ET
extraterminal
- fs(1)h
female sterile homeotic
- HPV
human papillomavirus
- PID
P-TEFb interacting domain
- P-TEFb
positive transcription elongation factor b
- PTD
protein transduction domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 2.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. J Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. J Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Florence B, Faller DV. Front Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 5.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Yik JH. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 8.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 9.Wu SY, Chiang CM. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 10.Jordan A, Bisgrove D, Verdin E. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. J Virol. 2006;80:9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPhillips MG, Ozato K, McBride AA. J Virol. 2005;79:8920–8932. doi: 10.1128/JVI.79.14.8920-8932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 14.Abbate EA, Voitenleitner C, Botchan MR. Mol Cell. 2006;24:877–889. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S. Mol Cell Biol. 2003;23:5354–5365. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gans M, Audit C, Masson M. Genetics. 1975;81:683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DH, Dawid IB. New Biol. 1990;2:163–170. [PubMed] [Google Scholar]
- 18.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]