Abstract
Aconitase (Aco1p) is a multifunctional protein: It is an enzyme of the tricarboxylic acid cycle. In animal cells, Aco1p also is a cytosolic protein binding to mRNAs to regulate iron metabolism. In yeast, Aco1p was identified as a component of mtDNA nucleoids. Here we show that yeast Aco1p protects mtDNA from excessive accumulation of point mutations and ssDNA breaks and suppresses reductive recombination of mtDNA. Aconitase binds to both ds- and ssDNA, with a preference for GC-containing sequences. Therefore, mitochondria are opportunistic organelles that seize proteins, such as metabolic enzymes, for construction of the nucleoid, an mtDNA maintenance/segregation apparatus.
Keywords: DNA binding, nucleoids, multifunction, mtDNA stability
Some proteins have evolved to have more than one function, with disparate activities displayed sometimes in different cellular compartments. Aconitase, an enzyme of the Krebs tricarboxylic acid (TCA) cycle localized in mitochondria, is a prototypical example of a multifunctional protein. In animals, aconitase also is found in the cytosol, in which it is known as the iron response protein 1 (IRP1). There, it reversibly binds to iron-responsive elements (IREs) in the 5′ and 3′ UTRs of mRNAs involved in iron metabolism, thereby regulating their translatability (1). Aconitase contains a 4[Fe–S] iron sulfur cluster (ISC), which is required for its enzymatic activity, but not for IRE–RNA binding. IRP1 has a hinge region separating protein domains 1–3 and 4 (2, 3). In response to iron depletion and the loss of ISC, the hinge region reversibly flexes to allow the two domains to open and close, providing a mechanism for clamping onto the stem loop structure in IREs.
In Saccharomyces cerevisiae, aconitase (Aco1p) also is a bifunctional protein. In addition to its TCA cycle function, Aco1p is a component of mtDNA nucleoids (4, 5), which are protein–DNA complexes associated with the inner mitochondrial membrane (6). These structures package mtDNA and otherwise assist in mtDNA transactions, including mtDNA inheritance. A number of nucleoid proteins have been identified in Xenopus lavis (7), mammals (8, 9), yeast (4, 5, 10, 11), and plants (12). Some have expected functions in mtDNA transactions, whereas others are well characterized metabolic proteins or chaperones, and hence their functions in nucleoids are unclear. Moreover, there is little overlap among these proteins in nucleoids from different species. Thus, if they are functional in nucleoids, their activities are either unique or different proteins fulfill similar functions in different organisms.
Yeast Aco1p mutants lose mtDNA (4). Like another bifunctional nucleoid protein, Ilv5p (13), the enzymatic and mtDNA maintenance functions of Aco1p are separable by mutation. For example, mutations in cysteine residues that coordinate to and thus attach the ISC to the protein result in a complete loss of Aco1p enzymatic activity but have only minor effects on mtDNA maintenance (4). An important feature of Aco1p is that its expression is under the control of two metabolic regulatory pathways: the heme activator protein (HAP) system, which is most active in cells with robust respiratory activity (14); and the retrograde (RTG) system, which is activated in cells with compromised or dysfunctional mitochondria (15, 16). When the HAP and RTG pathways are both inactivated, Aco1p is depleted and cells lose mtDNA (4). These findings suggest a mechanism involving Aco1p that couples energy metabolism to mtDNA maintenance (17). Here we find that Aco1p has an intrinsic affinity for both ds- and ssDNA, and that it potently protects mtDNA from damage. Thus, Aco1p provides an exceptional example of the successful evolution of a metabolic enzyme into a DNA- or RNA-binding protein that, depending on the species, carries out different biological activities in mitochondria and the cytosol.
Results
The Stability and Mutation Rate of ρ+ mtDNA Is Regulated by Aco1p Level.
mtDNA stability in yeast depends on Aco1p levels (Fig. 1 A and B). In rtg1Δ and rtg3Δ cells, which reduces ACO1 expression by ≈2-fold (Fig. 1A), the petite frequency significantly increases compared with wild-type cells (Fig. 1B). The increased mtDNA instability (e.g., in an rtg1Δ mutant strain) is largely reversed by ectopic expression of ACO1 from the RTG-independent ADH1 promoter. Moreover, spontaneous petite production in the wild-type strain can be reduced by the introduction of an additional copy of ACO1 and is completely suppressed by inactivation of MKS1, a negative regulator of the RTG pathway (18–20), which increases ACO1 expression by 4- to 6-fold (Fig. 1A). rtg1Δ, which abrogates ACO1 overexpression in mks1Δ cells, also abrogates the mtDNA stabilization effect of the mks1Δ allele. These results indicate a tight connection between Aco1p levels and mtDNA stability.
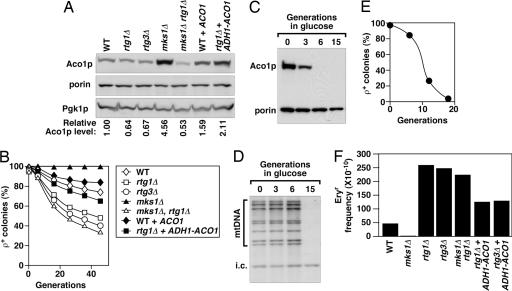
Fig. 1.
Aco1p levels are tightly correlated with the stability and mutation rate of ρ+ mtDNA. (A) Steady-state level of Aco1p. The level of porin is shown as a marker for mitochondrial proteins, and Aco1p levels relative to the wild-type control are normalized against the cytosolic Pgk1p. (B) Stability of the ρ+ genome in cells expressing different levels of Aco1p. mtDNA stability is expressed as a percentage of respiratory-competent (ρ+) colonies after growth in YPD for the number of generations as indicated. (C) Depletion of Aco1p in strain CS745–2C (aco1Δ::kan, ade2::GAL10-ACO1-URA3) during glucose repression in YPD medium. (D) Southern blot analysis of CfoI-digested total DNA probed for mtDNA in CS745–2C cells grown in YPD medium (i.c., internal control for sample loading). (E) Frequency of respiratory-competent cells in CS745–2C after growth in liquid YPD. The ρ+ cells were scored by the formation of red colonies when plated on YPGal medium. (F) Mutation rate in the ρ+ genome as expressed by the frequency of erythromycin-resistant (Eryr) colonies.
In cells where the only copy of ACO1 has been placed under the control of the glucose-repressible GAL10 promoter, the Aco1p level was reduced to undetectable levels after growth for six generations in glucose medium (Fig. 1C). However, at this time point, we did not observe any significant reduction in mtDNA copy number or any gross changes in the restriction profile of mtDNA (Fig. 1D). mtDNA was depleted to an undetectable level only after extended growth for 15 generations. In an independent assay, we measured the rate of petite formation during the shift from galactose to glucose medium. We found that >95% of the cells were converted to petites by 18 generations of growth in glucose medium (Fig. 1E). Petite production showed a distinct lag, which is consistent with the results of Fig. 1D. These findings suggest there are steps downstream of Aco1p that affect mtDNA maintenance and are deployed or inhibited when Aco1p synthesis is shut off.
The lack of apparent genome rearrangement immediately after Aco1p depletion in ρ+ cells invited the hypothesis that there might be progressive damage to mtDNA, such as the accumulation of point mutations, in response to Aco1p depletion. A complete loss of mtDNA might occur when these mutations accumulate and affect processes critical for ρ+ genome maintenance. To test this, we measured the frequency of mtDNA point mutations in cells expressing different levels of Aco1p by using the frequency of erythromycin-resistant (Eryr) colonies as a marker. Erythromycin resistance results from point mutations at positions 1951 and 3993 in the mitochondrial 21S rRNA gene (21, 22). This assay revealed that the mtDNA mutation rate is elevated by ≈5-fold in the rtg1Δ and rtg3Δ mutants with reduced ACO1 expression. Ectopic expression of ACO1 from the RTG-independent ADH1 promoter partially suppresses the mutator effect of both rtg1Δ and rtg3Δ, indicating that the down-regulation of ACO1 contributes to the increased mutation frequency. When Aco1p is overexpressed in an mks1Δ strain, the mutation rate is reduced by ≈20-fold. The antimutator effect of mks1Δ depends on the RTG pathway because an mks1Δ rtg1Δ double mutant has an mtDNA point mutation rate comparable to that of the rtg1Δ single mutant. Thus, there is a strong correlation between Aco1p levels (see Fig. 1A) and the rate of point mutations in mtDNA.
Aco1p Protects ρ− mtDNA from ssDNA Breaks and Reductive Genome Recombination.
Most genes affecting the stability of ρ+ mtDNA have little or no effect on the stability of ρ− petite mtDNAs (23–25). Petite mtDNAs are nonfunctional. Normally they are much less susceptible than ρ+ genomes to mutations in genes involved in mitochondrial transcription, translation, and repair activities, as well as to point mutations in their mtDNA (26). Nevertheless, Aco1p depletion leads to the rapid loss of ρ− mtDNA, as is shown for two ρ− petites, VAR1 and HS40, which contain repeated sequences of 1,800 and 760 bp of the VAR1 and ori5 genes, respectively (Fig. 2 A and B). The former encodes a mitochondrial ribosomal protein, whereas the latter encodes a putative origin of mtDNA replication. These data suggest that some fundamental aspects of mtDNA metabolism are compromised, which affects the stability of ρ− and ρ+ genomes, leading to the loss of mtDNA from both cell types in Aco1p-depleted cells. The relatively fast rate of the ρ− mtDNA loss versus its ρ+ counterpart may be explained by a counterselection against petites in the latter.
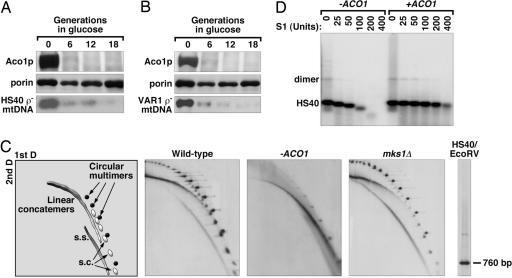
Fig. 2.
A role for Aco1p in the protection and maintenance of ρ− genomes. (A and B) Level of the HS40 and VAR1 ρ− mtDNA upon depletion of Aco1p in CS776/1 [aco1Δ, ade2::GAL10-ACO1-URA3, (HS40)] and CS777/2 [aco1Δ, ade2::GAL10-ACO1-URA3, (VAR1)] in glucose medium. Total DNA was digested with EcoRV and HincII and subjected to Southern blot by using probes specific for the HS40 and VAR1 ρ− genomes, respectively. Results of Western blot analysis are included to show the steady-state level of Aco1p and porin in the cells. (C) 2DAGE analysis of the HS40 ρ− mtDNA in cells depleted for Aco1p (−ACO1) or overexpressing ACO1 (mks1Δ), compared with cells expressing wild-type ACO1. (Left) Position of circular multimer, linear concatemer, supercoiled (s.c.), and the single-stranded (s.s.) forms of mtDNA. (Right) Position of the EcoRV-digested HS40 genome in the second-dimension electrophoresis. (D) S1 nuclease sensitivity of the HS40 ρ− genome extracted from cells with or without depletion of Aco1p.
We next followed possible changes to HS40 ρ− mtDNA during Aco1p depletion by 2D agarose gel electrophoresis (2DAGE) (Fig. 2C) (27). Compared with HS40 ρ− mtDNA from wild-type cells, HS40 mtDNA from cells partially depleted for Aco1p is dramatically downsized, with both the linear and circular forms of mtDNA redistributed to lower molecular weight species. By contrast, those forms of mtDNA from an mks1Δ mutant overexpressing ACO1 have an increased distribution of larger sized species, compared with those from the control strain. Aco1p thus regulates the size of the repeats of the ρ− genome.
Another important observation from the 2DAGE analysis is that the supercoiled forms of the HS40 circles are undetectable in cells partially depleted for Aco1p (Fig. 2C), suggesting an increase in the amount of ssDNA nicks in mtDNA. To test this, we assayed the sensitivity of mtDNA to S1 nuclease digestion in vitro, in which nicked sites as well as ssDNA are digested (Fig. 2D). We found that the HS40 ρ− mtDNA from the Aco1p-depeleted cells is significantly more sensitive to S1 nuclease digestion compared with mtDNA from the strain expressing ACO1. These observations suggest that Aco1p is likely required for protecting mtDNA from damage that promotes DNA nick/gap accumulation. The presence of excessive nicks may be accounted for by the genome downsizing, which is a result of abortive mtDNA replication and an increase in reductional recombination.
Accumulation of ssDNA Sites Destabilizes mtDNA in the Absence of Aco1p.
ssDNA is more vulnerable to mutations by DNA-damaging agents than dsDNA (28). We thus determined whether the presence of ssDNA sites in the mitochondrial genome correlates with the requirement for Aco1p in protecting mtDNA. One of the conditions that extensively increases the presence of damage-prone ssDNA regions is the formation of R-loop structures during or after transcription (29). ssDNA production is largely dependent on transcription in yeast mtDNA (30). The fact that transcription is not required for the maintenance of ρ− genomes (23) allowed us to test directly the hypothesis that blocking mitochondrial transcription may spare the requirement for Aco1p in mtDNA maintenance.
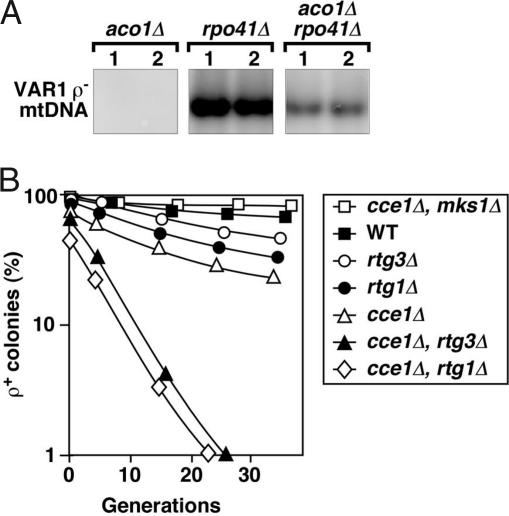
We therefore attempted to introduce the VAR1 ρ− genome into the aco1Δ and rpo41Δ single and aco1Δ rpo41Δ double mutants by cytoduction. RPO41 encodes the only known mitochondrial RNA polymerase activity essential for mitochondrial transcription (31). Cytoductants receiving VAR1 ρ− mtDNA were identified by direct colony hybridization. We were unable to identify any cytoductants carrying VAR1 in the aco1Δ background among >200 colonies analyzed, consistent with the essential role of Aco1p for the maintenance of ρ− genomes (Fig. 2B). By contrast, VAR1 can be successfully introduced into rpo41Δ cells with an efficiency of 50% (Fig. 3A). Interestingly, the VAR1 ρ− genome also can be cytoducted into the aco1Δ rpo41Δ recipient with a similar efficiency, in which it can be stably maintained despite a significantly reduced copy number, compared with what is seen in the rpo41Δ single mutant (Fig. 3A). These experiments indicate that loss of mitochondrial transcription can significantly stabilize mtDNA in cells lacking Aco1p, presumably by avoiding ssDNA site formation and thereby reducing a target of mtDNA damage.
Fig. 3.
mtDNA stability in Aco1p-depleted cells disrupted for RPO41 and CCE1. (A) Southern blot analysis showing the level of the VAR1 ρ− mtDNA in representative cytoductants with aco1Δ or rpo41Δ or aco1Δ plus rpo41Δ. (B) Frequency of respiratory-competent cells in cce1Δ mutants in which the RTG pathway was up- or down-regulated.
ssDNA sites also arise by DNA recombination intermediates. Thus, accumulation of Holliday junctions may represent another in vivo condition that would increase ssDNA regions. In yeast, CCE1 encodes a mitochondrial Holliday junction resolvase, and cce1Δ cells overaccumulate mtDNA recombination intermediates (32) and have unstable mtDNA in combination with defects in mtDNA base excision repair (33). A cce1Δ single mutant has a mildly increased frequency of petite production compared with the wild-type cells (Fig. 3B). We found that the integrity of the ρ+ genome is dramatically compromised when combining the cce1Δ allele with rtg1Δ or rtg3Δ, which reduces ACO1 expression (see Fig. 1A). By contrast, mks1Δ significantly suppresses petite formation in cce1Δ cells. These observations lend further support for a synergistic effect on mtDNA instability between reduced Aco1p activity and mtDNA transactions involving the accumulation of ssDNA sites that would be susceptible to damage.
Epistasis with the mtDNA Repair/Recombination Helicase, Pif1p.
We next examined possible genetic interactions between ACO1 and genes directly involved in processes such as the mutagenesis and repair of mtDNA [see supporting information (SI) Text]. We found a strong epistasis with PIF1 by encoding a DNA helicase involved in mtDNA repair and recombination (34). pif1 mutants have an unstable mitochondrial genome, which is further destabilized by a compromise in mtDNA base excision repair (33). Pif1p also is required to maintain mtDNA under conditions that induce dsDNA breaks, suggesting that it has a role in the repair or prevention of dsDNA breaks (35). Importantly, like Aco1p and Abf2p, Pif1p is associated with mtDNA as a nucleoid protein.
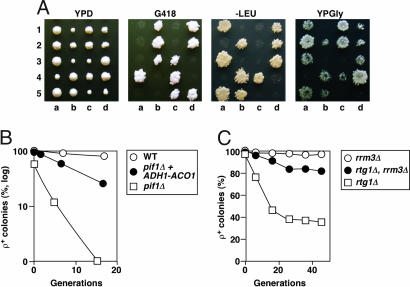
We found that mtDNA instability in pif1Δ cells is suppressed by mks1Δ. By meiotic segregational analysis of pif1Δ mks1Δ heterozygous diploids (Fig. 4A), the nascent pif1Δ segregants (G418R) form respiratory-deficient colonies on glucose medium as revealed by their inability to grow on glycerol medium because of the loss of mtDNA. However, all of the pif1Δ mks1Δ double mutants (G418R, Leu+) among the segregants form colonies after replica plating on glycerol medium, suggesting that mks1Δ suppresses mtDNA instability in pif1Δ cells. Importantly, mtDNA instability in the pif1Δ mutant can be suppressed by overexpression of ACO1 from an ADH1 promoter (Fig. 4B). Furthermore, disruption of RRM3, a gene that suppresses mtDNA instability in pif1 mutants by activating the Mec1/Rad53 nuclear checkpoint pathway and regulating deoxyribonucleoside triphosphate pools (36), also suppresses mtDNA loss in the rtg1Δ mutant that has reduced ACO1 expression (Fig. 4C). Therefore, Aco1p is epistatic to Pif1p in mtDNA maintenance.
Fig. 4.
Epistasis between ACO1 expression and PIF1. (A) Growth phenotype of five complete tetrads from CS956 (pif1Δ::kan/+, mks1Δ::LEU2/+). (B) Frequency of respiratory-competent cells after growth in YPD medium. (C) Suppression of mtDNA instability in rtg1Δ mutant by rrm3Δ.
Aco1p Has an Intrinsic DNA-Binding Activity.
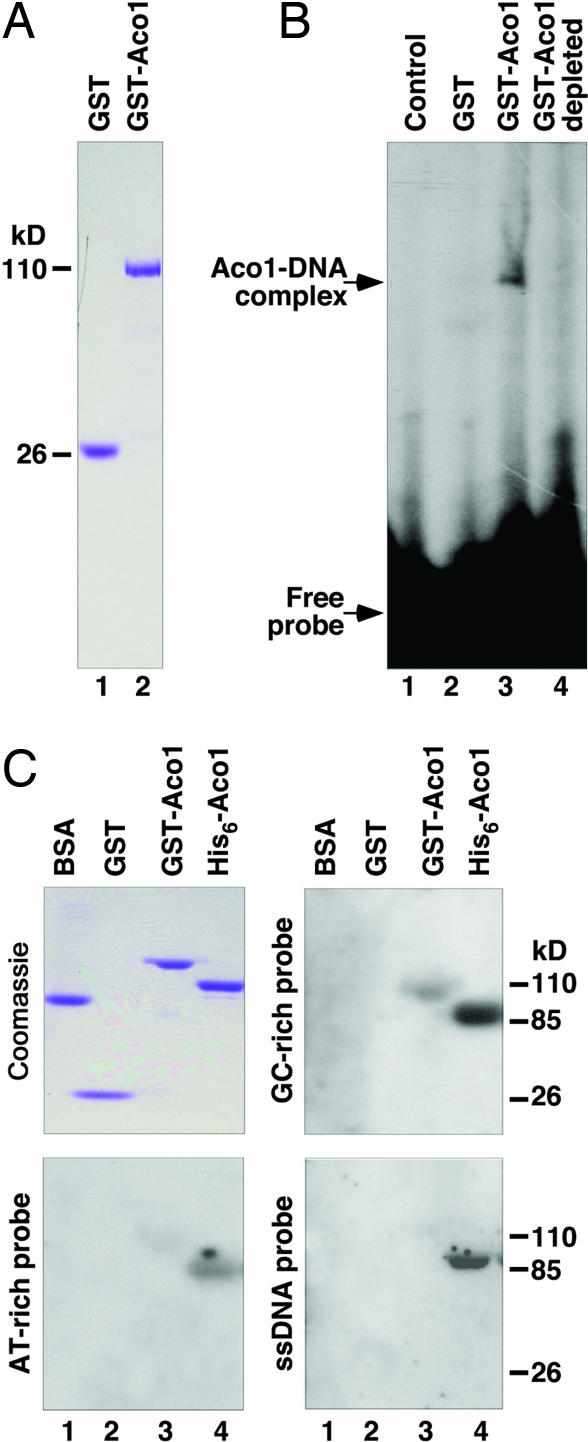
Overexpression of ACO1 suppresses mtDNA instability in cells lacking the key mtDNA packaging factor, Abf2p (4). A parsimonious view is that, like Abf2p, Aco1p binds directly to dsDNA, protecting it from the overall accumulation of damages targeted to various vulnerable sites. To test this finding, we overexpressed Aco1p tagged at its N terminus with GST and purified the fusion protein from Escherichia coli (Fig. 5A). By gel mobility shift assays, GST-Aco1p forms a discrete DNA–protein complex with a dsDNA probe (Fig. 5B). The fact that only a small portion of the probe is shifted in its mobility suggests that GST-Aco1p has weak DNA-binding activity.
Fig. 5.
Aco1p binds to DNA. (A) Coomassie-stained gel showing purified GST and GST-Aco1p. (B) Gel retardation assay showing the binding of GST-Aco1 to the AT-rich dsDNA probe, HS40ATFR (SI Text). Lane 1, no protein; lane 2, GST (2 μg); lane 3, GST-Aco1p (2 μg); lane 4, equal volume of aliquot after GST-Aco1p depletion by glutathione Sepharose 4B beads. (C) Coomassie-stained gel showing purified GST-Aco1p and His6-Aco1p, and Southwestern blot showing the binding of the fusion proteins to GC- and AT-rich ds- and ssDNA.
We confirmed the DNA-binding ability of GST-Aco1p by Southwestern blot assay (Fig. 5C). Using the same assay, we found that the N-terminal His6-tagged form of Aco1p binds dsDNA with a significantly higher affinity compared with GST-Aco1p likely because of improved solubility or renaturation efficiency of His6-Aco1p compared with GST-Aco1p. Both GST-Aco1p and His6-Aco1p have a significantly lower affinity toward an AT-rich probe. The two fusion proteins also are capable of binding to an ssDNA probe, with His6-Aco1p having a much stronger binding affinity compared with GST-Aco1p.
DNA-Binding Activity Is Required for mtDNA Maintenance.
Is the DNA-binding activity of Aco1p required for mtDNA maintenance? To address this question, we looked for mutations that compromise Aco1p's ability to bind to DNA in vitro and to maintain mtDNA in vivo. We assumed that Aco1p, like IRP1 in animals, also may use the cleft between domains 1–3 and 4 for interacting with DNA. To test this, we generated the R476S and R668S mutants of Aco1p, equivalent to Arg-541 and Arg-780 in IRP1, which are located on the docking surface for IRE–RNA after the opening of the binding cleft between domains 3 and 4 (see Fig. 6A) (2, 3). Arg-541 of IRP1 stabilizes the interaction with the RNA terminal loop through a network of hydrogen bonds, and Arg-780 directly interacts with a cytidine on the IRE stem. Mutations of those residues in IRP1 diminish but do not completely abolish the binding to IRE–RNA (37, 38).
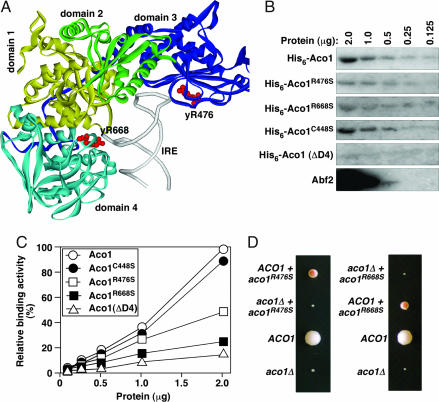
Fig. 6.
Aco1p variants compromised in DNA-binding in vitro are defective in mtDNA maintenance in vivo. (A) The position of Arg-541 and Arg-780 in rabbit IRP1 (equivalent to yArg476 and yArg668) in the IRE-binding conformation (3). (B) Southwestern blot showing the binding of Aco1p variants and Abf2 to the GC-rich BA box probe. Abf2p is loaded at a concentration 20-fold lower than the Aco1p variants. (C) Relative affinity of Aco1p variants to the GC-rich B and A box probe. (D) Representative tetrads showing that the meiotic segregants expressing only aco1R476S or aco1R668S form petite colonies on complete raffinose medium.
We found that the binding of His6-Aco1pR476S and His6-Aco1pR668S to dsDNA is reduced by 2.0- and 3.8-fold, respectively, compared with His6-Aco1p (2 μg of the proteins loaded) (see Fig. 6 B and C). Arg-668 is located on the domain 4 side of the interdomain cleft. A truncated version of the protein, which lacks the entire domain 4 (Aco1p-ΔD4), has barely detectable DNA-binding activity (Fig. 6 B and C), suggesting that domain 4 is important for interacting with DNA. Under these conditions, His6-Aco1p binds to dsDNA with an efficiency that is much lower than His6-Abf2p.
We finally introduced the aco1R476S and aco1R668S alleles into a diploid strain heterozygous for aco1Δ by chromosomal integration. After sporulation and tetrad dissection, meiotic segregants were identified that express only aco1R476S or aco1R668S (Fig. 6D). Like aco1Δ segregants, the aco1R476S and aco1R668S cells form small colonies that completely lack mtDNA (data not shown). Thus, aco1R476S and aco1R668S are unable to maintain mtDNA in vivo. Overexpression of aco1R476S and aco1R668S from the ADH1 promoter could partially rescue cells from mtDNA loss (data not shown). However, the aco1C448S variant, which abrogates the ISC ligation and the catalytic activity of Aco1p, but is still active in mtDNA maintenance (4), largely retains the DNA-binding activity of the protein (Fig. 6 B and C). Taken together, our data suggest that a discrete DNA-binding activity is required for Aco1p to protect and maintain mtDNA. Despite the fact that no DNA-binding activity has been found to be associated with IRP1, at least some common residues are likely used by Aco1p and IRP1 to interact with DNA and RNA, respectively.
Discussion
Our findings have established a possible mechanism for yeast Aco1p in mtDNA maintenance. Cells depleted of Aco1p suffer an increase in point mutations and accumulate ssDNA nicks in mtDNA. These events may lead to replication stalling and reductive recombination, which are consequently manifested by progressive size contraction and, ultimately, complete loss of mitochondrial genomes. Our data suggest a role of Aco1p in protecting the mitochondrial genome from damage through direct binding to mtDNA. Although the details of Aco1p–DNA interactions remain to be elucidated, the binding activity to dsDNA provides an explanation for the suppression of mtDNA instability by ACO1 overexpression in cells lacking the mtDNA packaging factor, Abf2p (4). Therefore, Aco1p and Abf2p can recognize dsDNA as a common substrate. Like Abf2p, Aco1p also shows preferential binding to GC-containing sequences in vitro. Because Aco1p is essential for mtDNA maintenance even in the presence of Abf2p, the functional activities of Aco1p and Abf2p do not completely overlap. Because Aco1p also binds to ssDNA, we speculate that Aco1p, but not Abf2p, may protect ssDNA sites.
Given the DNA-damaging effects of Aco1p depletion, it is perhaps not surprising that ρ− petite mitochondrial genomes also are unstable in Aco1p-depleted cells. This finding is a rare example of how a gene affects the stability of both ρ+ and ρ− mtDNAs except for those directly involved in mtDNA replication. The finding also substantiates the conclusion that Aco1p is stabilizing mtDNA not through redox routes, because ρ− petite cells are respiratory-deficient, but through control of primary DNA activities, such as replication, recombination, or repair. This mtDNA maintenance system is poised with great sensitivity in that a ≈2-fold reduction of in vitro DNA-binding activity of Aco1p in some mutants is sufficient to cause mtDNA instability in vivo. This sensitivity corroborates the significant suppression of mtDNA instability in abf2Δ mutants by a 2-fold increase in ACO1 expression (4). Whereas mutants of Aco1p that lack enzymatic activity, such as those lacking the ISC, can still maintain mtDNA, these findings do not mean that mtDNA stability is no longer linked to the metabolic state of the cell. On the contrary, Aco1p expression can be up-regulated by 2- to 4-fold in response to respiratory growth or to mitochondrial dysfunction through the HAP and RTG regulatory pathways. Under these conditions, mtDNA transactions such as transcription and recombination are actively induced, concomitant with an increase in ssDNA sites and the production of free radicals (39). The up-regulation of Aco1p could play a critical role in providing an adequate protection to ss- and dsDNA. Future studies will be directed toward investigating the relative distribution of Aco1p between nucleoids and the matrix and understanding whether Aco1p is present in the nucleoids in the apoprotein or holoprotein forms.
The mtDNA nucleoid is a unique structure in which many of the proteins found there have been recruited to fulfill functions estranged from their “normal” or primary activities (17). For instance, in addition to the DNA-binding Aco1p, the yeast NAD+-specific isocitrate dehydrogenase, which is a nucleoid component, also binds to mRNA (40). This result raises the remarkable possibility that mtDNA nucleoids contain the highest density of multifunctional proteins within the cell. Those functions likely represent secondary evolutionary events given that the primary activities of these proteins are conserved among species. The nucleoid activities are, for the most part, unique to different species. Animal cells, for example, rely largely on glucose as a source of energy and tailor their metabolic activities to that metabolic fact. In contrast, yeast strains switch between fermentative and oxidative metabolism as part of their normal growth behavior, which influences mitochondrial gene expression in fundamentally different ways than in animal cells. Therefore, it is reasonable to expect that these different environmental cues and responses are reflected by differences in the composition of nucleoids, making mitochondria opportunistic in the recruitment and utilization of diverse proteins required for nucleoid function. This also drives the successful evolution of aconitase into a DNA-binding protein in yeast mitochondria, in parallel to its evolution into the RNA-binding IRP1 in animals.
Materials and Methods
Growth Media and Yeast Strains.
Complete yeast extract/peptone/dextrose (YPD) and yeast nitrogen base minimal medium were prepared with 2% dextrose, 2% galactose, or 2% glycerol (Gly). All of the yeast strains used in this study are isogenic to S288C (see SI Table 1). Yeast strains and plasmids were constructed as described in SI Text.
Estimation of the mtDNA Mutation Rate.
The mtDNA mutation rate was estimated based on the erythromycin-resistance assay as described by Chi and Kolodner (41). Twenty to 25 individual colonies for each strain from yeast extract/peptone (YP)Gly plates were inoculated into 5 ml of YPD medium and were grown at 30°C for 48 h to an optical density of 2 to 4 at 600 nm. Aliquots of the cultures were diluted for enumerating respiring cells on YPGly plates. The remainder of the cells were plated onto solid YPGly medium containing erythromycin (1 mg/ml). Erythromycin-resistant colonies were scored after incubating at 30°C for 9 days. The erythromycin-resistant mutation rate was calculated by using the formula μ = −ln(P(0))/n × ln 2 and expressed as mutation per cell per generation.
2DAGE and S1 Nuclease Sensitivity Assay.
2DAGE was carried out according to MacAlpine et al. (30). Details are described in SI Text. For S1 nuclease sensitivity assay, total DNA was extracted from cells expressing or depleted of Aco1p. Five micrograms of DNA was digested with DraI, which linearizes the HS40 ρ− genome, and then treated with S1 nuclease (Roche, Indianapolis, IN) in 30 mM Na-Acetate (pH 4.6), 1 mM ZnSO4, and 5% Gly at 37°C for 30 min. The reactions were terminated by 10 mM EDTA (pH 8.0) before being subjected to Southern blot analysis.
Gel Retardation Assay and Southwestern Analysis.
GST- and His6-tagged Aco1p variants were produced in E. coli and used for gel retardation assay and Southwestern analysis as described in SI Text. The GC-containing B and A box probe used for DNA binding is HS40BAF (5′-GAAGATATCCGGGTCCCAATAATAATTATTATTGAAAATAATAATTGGGACCCCCACAATAAA-3′, with the B and A boxes in italic type). The AT-rich probe is HS40ATF (5′-TTAATATTTAATAATATAATCAATAAATAATATTATAATAATATAATATAA-3′). The probes were used in single- or double-stranded forms. Both sequences are derived from the HS40 ρ− genome.
Supplementary Material
Acknowledgments
We thank A. Tizenor for help with the graphics and our colleagues for helpful discussions and a critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (to X.J.C. and R.A.B.) and a grant from the Robert A. Welch Foundation (to R.A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703078104/DC1.
References
- 1.Rouault TA. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 2.Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, Fontecilla-Camps JC. Structure (London) 2006;14:129–139. doi: 10.1016/j.str.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 4.Chen XJ, Wang X, Kaufman BA, Butow RA. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman BA, Newman SM, Hallberg RL, Slaughter CA, Perlman PS, Butow RA. Proc Natl Acad Sci USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albring M, Griffith J, Attardi G. Proc Natl Acad Sci USA. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Mol Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Bogenhagen DF. J Biol Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 10.Meeusen S, Tieu Q, Wong E, Weiss E, Schieltz D, Yates JR, Nunnari J. J Cell Biol. 1999;145:291–304. doi: 10.1083/jcb.145.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyakawa I, Sando N, Kawano S, Nakamura S, Kuroiwa T. J Cell Sci. 1987;88:431–439. doi: 10.1242/jcs.88.4.431. [DOI] [PubMed] [Google Scholar]
- 12.Dai H, Lo YS, Litvinchuk A, Wang YT, Jane WN, Hsiao LJ, Chiang KS. Nucleic Acids Res. 2005;33:4725–4739. doi: 10.1093/nar/gki783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman JM, Perlman PS, Butow RA. Genetics. 2002;161:1043–1052. doi: 10.1093/genetics/161.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeRisi JL, Iyer VR, Brown PO. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 15.Butow RA, Avadhani NG. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Butow RA. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 17.Chen XJ, Butow RA. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 18.Dilova I, Chen CY, Powers T. Curr Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- 19.Sekito T, Liu Z, Thornton J, Butow RA. Mol Biol Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tate JJ, Cox KH, Rai R, Cooper TG. J Biol Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sor F, Fukuhara H. Nucleic Acids Res. 1982;10:6571–6577. doi: 10.1093/nar/10.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sor F, Fukuhara H. Nucleic Acids Res. 1984;12:8313–8318. doi: 10.1093/nar/12.22.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fangman WL, Henly JW, Brewer BJ. Mol Cell Biol. 1990;10:10–15. doi: 10.1128/mcb.10.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelenaya-Troitskaya O, Newman SM, Okamoto K, Perlman PS, Butow RA. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo XM, Clark-Walker GD, Chen XJ. Genetics. 2002;160:1389–1400. doi: 10.1093/genetics/160.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Contamine V, Picard M. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brewer BJ, Fangman WL. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl T, Nyberg B. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Manley JL. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 30.MacAlpine DM, Kolesar J, Okamoto K, Butow RA, Perlman PS. EMBO J. 2001;20:1807–1817. doi: 10.1093/emboj/20.7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenleaf AL, Kelly JL, Lehman IR. Proc Natl Acad Sci USA. 1986;83:3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lockshon D, Zweifel SG, Freeman-Cook LL, Lorimer HE, Brewer BJ, Fangman WL. Cell. 1995;81:947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 33.O'Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mol Cell Biol. 2002;22:4086–4093. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng X, Dunaway S, Ivessa AS. Mitochondrion. 2006;7:211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 36.Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O'Rourke T, Siede W, Shadel GS. Mol Biol Cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butt J, Kim HY, Basilion JP, Cohen S, Iwai K, Philpott CC, Altschul S, Klausner RD, Rouault TA. Proc Natl Acad Sci USA. 1996;93:4345–4349. doi: 10.1073/pnas.93.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philpott CC, Klausner RD, Rouault TA. Proc Natl Acad Sci USA. 1994;91:7321–7325. doi: 10.1073/pnas.91.15.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barros MH, Netto LE, Kowaltowski AJ. Free Radical Biol Med. 2003;35:179–188. doi: 10.1016/s0891-5849(03)00307-1. [DOI] [PubMed] [Google Scholar]
- 40.Anderson SL, Schirf V, McAlister-Henn L. Biochemistry. 2002;41:7065–7073. doi: 10.1021/bi0200662. [DOI] [PubMed] [Google Scholar]
- 41.Chi NW, Kolodner RD. J Biol Chem. 1994;269:29984–29992. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.