Abstract
The two primary human inflammatory bowel diseases, Crohn's disease (CD) and ulcerative colitis (UC), are idiopathic relapsing disorders characterized by chronic inflammation of the intestinal tract. Although several lines of reasoning suggest that gastrointestinal (GI) microbes influence inflammatory bowel disease (IBD) pathogenesis, the types of microbes involved have not been adequately described. Here we report the results of a culture-independent rRNA sequence analysis of GI tissue samples obtained from CD and UC patients, as well as non-IBD controls. Specimens were obtained through surgery from a variety of intestinal sites and included both pathologically normal and abnormal states. Our results provide comprehensive molecular-based analysis of the microbiota of the human small intestine. Comparison of clone libraries reveals statistically significant differences between the microbiotas of CD and UC patients and those of non-IBD controls. Significantly, our results indicate that a subset of CD and UC samples contained abnormal GI microbiotas, characterized by depletion of commensal bacteria, notably members of the phyla Firmicutes and Bacteroidetes. Patient stratification by GI microbiota provides further evidence that CD represents a spectrum of disease states and suggests that treatment of some forms of IBD may be facilitated by redress of the detected microbiological imbalances.
Keywords: Crohn's disease, culture-independent microbiology, ulcerative colitis, rRNA
The human inflammatory bowel diseases (IBD), ulcerative colitis (UC) and Crohn's disease (CD), are chronic gastrointestinal (GI) illnesses of uncertain etiology, with symptoms ranging from diarrhea and weight loss to ulceration, perforation, and complete obstruction of the GI tract (1–3). Both diseases are incurable and often require extensive medical and surgical management. Medical treatment of IBD with antibiotic, antiinflammatory, and/or immunosuppressive drugs is generally supportive rather than curative (4). Clearly, novel approaches to disease management are needed and will require more thorough understanding of the factors that contribute to IBD.
IBD is manifest in the context of an immense and highly diverse population of commensal GI microbes. Studies of experimental animal models of IBD reveal that germ-free animals normally manifest few signs of inflammation (5–14); the full effects of experimental colitis typically are exhibited only upon exposure to natural microbial communities (9, 12, 13). A role for microbes in human IBD similarly is suggested by the observation that fecal flow exacerbates inflammation of involved tissues (15–18). Current models of human IBD posit that pathogenesis arises from, and is perpetuated by, interactions between host genetic and immune factors, GI microbes, and environmental triggers (8). The net effect is a dysregulated hypersensitive TH1-type response of the gut-associated lymphoid tissue to luminal microbes and/or their antigens (2, 8). Considerable controversy remains, however, as to whether the entire commensal microbiota or individual pathogens [e.g., Mycobacterium avium spp. paratuberculosis (MAP)] are primarily responsible for induction of inflammation (19–24).
The commensal GI microbiota provides a multitude of beneficial services to the healthy host, including maintenance of immune homeostasis (25–27), modulation of GI development (28, 29), and enhanced metabolic capabilities (30). Perturbations of the normally stable GI microbiota might therefore be predicted to adversely affect the health of the host. Indeed, recent studies demonstrate that obesity in humans and ob/ob mice is associated with stereotypical imbalances in the normal gut microbiota (31–33). Likewise, previous studies of human IBD, using standard culture techniques (34–36) or molecular analysis (37–43), have noted alterations in the GI microbiota. However, most of these IBD studies have been limited in statistical power and precision of identification or have examined only the fecal microbiota, which differs substantially from that of the GI mucosa (44). Because of their immediate proximity to affected tissues, microbes associated with the gut wall likely are a more critical factor than are fecal microbes in promoting IBD pathogenesis (45, 46). These previous studies also have treated UC and CD as monolithic disorders, thus precluding the possibility that different subpopulations exist within IBD classifications (47, 48). To surmount these problems, we performed a comprehensive culture-independent phylogenetic analysis of microbes associated with a greatly expanded set of surgically resected tissue specimens. Our findings indicate significant abnormalities in the microbiotas of a subset of CD and UC individuals.
Results
Broad-Range PCR Analysis.
One hundred ninety resected tissue samples were obtained in approximately equal numbers from CD, UC, and non-IBD control subjects (primarily individuals being treated for GI cancer). The clinical features of the specimen set are summarized in supporting information (SI) Table 2. To minimize the presence of luminal bacteria, tissues were rinsed in 0.15 M NaCl before extraction of total genomic DNA. Fifteen thousand one hundred and seventy-two small-subunit rRNA (SSU rRNA) genes (≈80 per sample) were cloned and sequenced from these samples after broad-range PCR with panbacterial primers. The kinds of microbes resident in the resected tissues were identified by molecular-phylogenetic sequence analysis. Few identical sequences were observed between different samples, and so similar sequences were clustered into operational taxonomic units (OTUs). OTUs were assembled at a range of sequence identity thresholds so that sample sets could be compared at multiple phylogenetic depths (SI Fig. 6). No significant differences in sampling coverage were evident between sample categories at any OTU clustering level (SI Table 3), thus permitting meaningful comparison of samples.
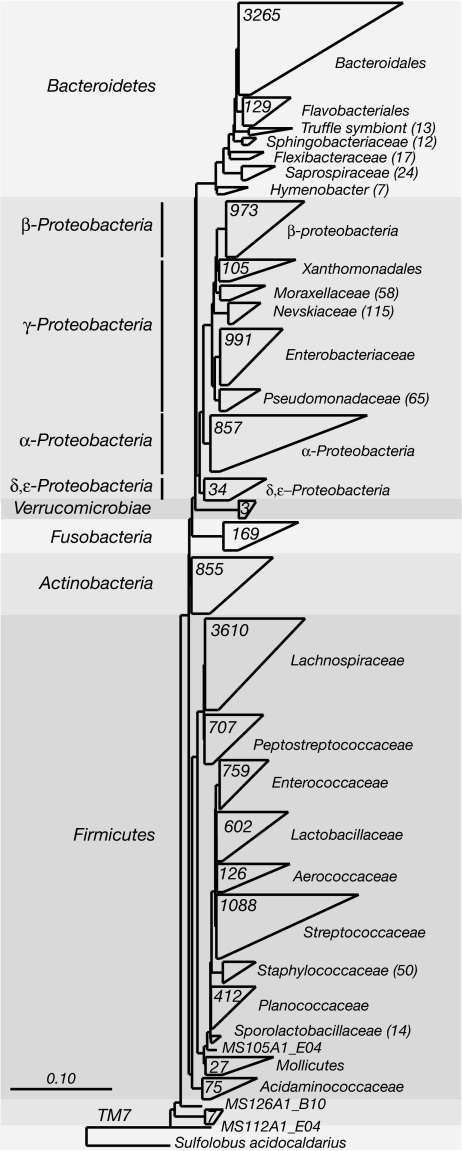
The overall phylogenetic distribution of the sequences identified in this study is shown in Fig. 1. Regardless of disease state or anatomical site of sampling, the majority of sequences were associated with only four phyla of the Bacteria (Figs. 1 and 2): Firmicutes (49% of clones), Bacteroidetes (23%), Proteobacteria (21%), and Actinobacteria (5%). Nearly half of the sequences (≈45%) belonged to just two subgroups: the order Bacteroidales and the family Lachnospiraceae (which comprises the Clostridium XIVa and IV groups within the order Clostridiales; ref. 49).
Fig. 1.
Phylogenetic distribution of SSU rRNA sequences. The diagrammatic phylogenetic tree presents a summary of the rRNA sequences isolated in this study. The number of taxa in a clade is designated either on its wedge or parenthetically to its name. Phyla are named to the left of the tree and lower taxonomic levels to the right. Wedge widths represent the taxa with the longest (top bar) and shortest (bottom bar) distances within the clade. The scale bar represents base changes per site.
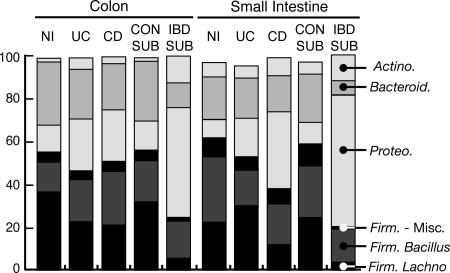
Fig. 2.
Phylum-level comparison of disease state. Bars shading depicts percentages of cloned sequences in all samples of a particular category that belong to one of the predominant phylum of Bacteria identified in the sample set (Firmicutes are subdivided into Bacillus, Lachnospiraceae, and miscellaneous groups). Phyla of lower abundance were omitted, so bars do not sum to 100%. NI, non-IBD control samples; Con-Sub, Control subset; and IBD-Sub, IBD subset are explained in the text.
Compared with the colon, samples of the small intestine were substantially enriched in sequences of the Bacillus subgroup of Firmicutes (primarily Streptococcaceae). In the case of non-IBD control samples, Streptococcaceae sequences accounted for 23% and 5% of total clones isolated, respectively, from the small intestine and colon (Fig. 2 and SI Fig. 7). Actinobacterial sequences also were more prevalent in the small intestine (8% of clones in non-IBD samples compared with 2% from non-IBD colon). Members of the actinobacterial subgroups Actinomycinaeae and Corynebacteriaceae accounted for the majority of this enrichment. In contrast, small-intestine samples contained fewer sequences of Bacteroidetes and Lachnospiraceae, compared with colonic samples.
OTU distributions were significantly associated with disease state (P = 0.02) but not anatomy (P = 0.39) or gross pathology (P = 0.25). Relative to non-IBD control colon samples, UC and CD colonic samples contained fewer sequence types representative of Bacteroidetes and the Lachnospiraceae subgroup of Firmicutes and concomitantly more sequences representative of Proteobacteria and the Bacillus subgroup of Firmicutes (Fig. 2). Among the small-intestine samples, proteobacterial sequences were more abundant and Bacillus sequences less abundant in both UC and CD sample sets relative to non-IBD samples. Lachnospiraceae sequences of non-IBD samples were intermediate in abundance between UC and CD samples, whereas Bacteroidetes sequences were unaffected by disease status. Similar results were observed when clonal prevalences, rather than frequencies, were compared across sample sets (SI Fig. 8).
Exploratory Data Analysis.
Because of the idiopathic nature of IBD, we conducted exploratory statistical analyses to determine whether any structure is manifest within the sequence data set, independently of diagnostic categories. Principal components analyses (PCA) of sample vs. OTU presence/absence matrices were performed for OTUs defined at several phylogenetic depths (97% OTU results are shown in Fig. 3). MANOVA tests of the 97% OTU PCA data indicated that variation along the first two principal coordinate axes was significantly influenced by disease status (P < 0.001) rather than anatomy (P = 0.4) or pathology (P = 0.7). Similar results also obtained for PCA of OTUs defined at the 99%, 95%, 90%, and 85% levels (SI Fig. 9 and SI Table 4).
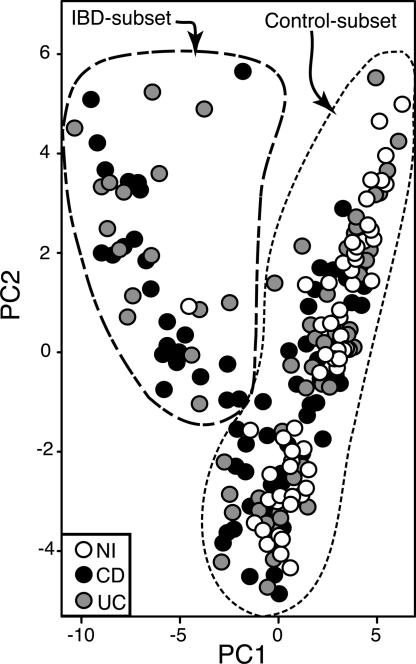
Fig. 3.
PCA; 97% OTUs were encoded as presence/absence data for each sample, and the corresponding data matrix was subjected to PCA. Each circle, which is representative of a single sample and shaded according to disease status, is plotted along the first two principal component axes. Dashed lines denote nominal sample clusters, the Control subset and IBD subset.
Hierarchical clustering of samples (SI Fig. 10) on the basis of their first two principal component scores separated the samples into two primary clusters (outlined in Fig. 3). One cluster (labeled “Control subset”) includes most of the non-IBD control samples as well as ≈2/3 of CD and ≈3/4 of UC samples. The other nominal cluster is dominated by CD and UC samples and is thus designated the “IBD subset.” Hierarchical clustering of samples by raw OTU presence/absence produced similar results (SI Fig. 11), although more IBD samples clustered with the IBD subset. Furthermore, the UniFrac test (50), which assesses the significance of intercommunity differences in populations, strongly supports our hypothesis that IBD subset and Control subset specimens are composed of distinct microbial communities (SI Fig. 12).
The differences in rRNA clonal frequency that are evident from comparisons of disease state are even more pronounced when the data are parsed in terms of inclusion in, or exclusion from, the IBD subset (Fig. 2). Sequences representative of the Bacteroidetes (P < 0.001) and Lachnospiraceae (P < 0.001) were greatly depleted, and those of the Actinobacteria (P < 0.001) and Proteobacteria (P < 0.001) were substantially more abundant in IBD-subset samples, relative to Control-subset samples. Summaries of the 97% OTU clusters that were the most depleted or enriched in prevalence in the IBD-subset samples are presented in Table 1(more detailed lists are available in SI Tables 5 and 6). A diverse variety of Actinobacteria and Proteobacteria was increased in prevalence in the IBD subset. In contrast, most of the OTUs that were decreased in prevalence in the IBD subset belonged to either the Lachnospiraceae group or the phylum Bacteroidetes.
Table 1.
Ninety-seven percent OTUs enriched or depleted in IBD subset
| Top Blast hit* | Phylum† | ΔPrevalence,‡% |
|---|---|---|
| Depleted in IBD subset | ||
| Bacterium mpn-isolate group 5 | Bacteroidetes; Bacteroidales | −42.3 |
| Bacterium mpn-isolate group 18 | Firmicutes; Lachnospiraceae | −37.7 |
| Butyrate-producing bacterium A2–A165 | Firmicutes; Lachnospiraceae | −34.0 |
| Butyrate-producing bacterium SR1/1 | Firmicutes; Lachnospiraceae | −32.5 |
| Bacteroides thetaiotaomicron | Bacteroidetes; Bacteroidales | −32.5 |
| Butyrate-producing bacterium L2—L7 | Firmicutes; Lachnospiraceae | −27.6 |
| Clostridium nexile | Firmicutes; Lachnospiraceae | −26.2 |
| Bacterium mpn-isolate group 19 | Firmicutes; Lachnospiraceae | −26.2 |
| Butyrate-producing bacterium SS2/1 | Firmicutes; Lachnospiraceae | −24.1 |
| Alistipes sp. WAL 8169 | Bacteroidetes; Bacteroidales | −24.0 |
| Enriched in IBD subset | ||
| Drinking-water bacterium Y7 | Alphaproteobacteria | 76.0 |
| Actinobacterium GWS-BW-H99 | Actinobacteria | 62.5 |
| Nocardioides sp. NS/27 | Actinobacteria | 61.8 |
| Novosphingobium sp. K39 | Alphaproteobacteria | 57.8 |
| Pseudomonas straminea | Betaproteobacteria | 53.7 |
| Gamma proteobacterium DD103 | Gammaproteobacteria | 50.9 |
| Bacillus licheniformis | Firmicutes;Bacilli | 49.7 |
| Sphingomonas sp. AO1 | Alphaproteobacteria | 49.6 |
| Actinomyces oxydans | Actinobacteria | 48.9 |
| Acidimicrobidae Ellin7143 | Actinobacteria | 47.6 |
*Identified by Blast search of database culled of environmental clones.
†Determined by comparison to rRNA sequence database (49).
‡Difference in prevalence of an OTU between normal subset and IBD subset.
The disparities in microbiotas noted between the nominal IBD and Control subsets might simply be explained in terms of different medical treatments to which patients were subjected. However, none of the treatments listed in patient records were significantly correlated with classification of samples into the IBD or Control subset, when analyzed individually or in toto (SI Table 7). In terms of reported disease complications, only the occurrence of abscesses in CD subjects was strongly predictive of IBD-subset inclusion: 8/24 IBD-subset CD samples and 0/44 Control-subset CD samples were positive for abscesses (P < 0.001; SI Table 7). CD subjects with a history of smoking were significantly more likely to be classified as Control subset than were reported nonsmokers (P = 0.007). Patient age at time of surgery also was correlated with the IBD/Control-subset split (SI Table 8); members of the IBD subset were, on average, 9.5 (CD; P = 0.014) or 6.3 (UC; P = 0.08) years younger than Control-subset subjects. Sex and previous appendectomy were not correlated with GI microbiota composition.
Quantification of Specific Microbial Groups.
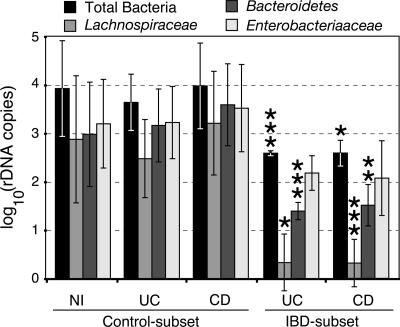
Shifts in OTU frequency in clone libraries could result from either blooms or diminished loads of particular microbial groups. To distinguish these possibilities, quantitative PCR (Q-PCR) analyses of total bacteria, Bacteroidetes, Enterobacteriaceae (a proxy for Proteobacteria), and Lachnospiraceae were performed for selected IBD- and Control-subset samples. As shown in Fig. 4, Q-PCR results revealed a 10-fold decrease in total bacterial load in the IBD subset (P < 0.001). Q-PCR also confirmed that members of the Lachnospiraceae (P < 0.001) and Bacteroidetes (P < 0.001) were diminished in quantity in the IBD-subset samples (statistical analyses were performed on data that were normalized to the Q-PCR measurements of total bacteria). The Q-PCR results indicated >300-fold (Lachnospiraceae) and 50-fold (Bacteroidetes) reductions in these microbial groups in the IBD subset, compared with the Control subset. In contrast, the measured levels of Enterobacteriaceae were not substantially different in IBD- and Control-subset samples (P = 0.47). The differences in IBD- and Control-subset clonal frequencies must therefore reflect diminution of Lachnospiraceae and Bacteroidetes microbial populations in patients' samples classified as IBD subset rather than increased absolute levels of proteobacteria.
Fig. 4.
Q-PCR analysis of predominant phylogenetic groups. Selected representatives of the Control- and IBD-subset clusters (see text for details) were assayed by Q-PCR for total bacteria, Lachnospiraceae, Bacteroidetes, and Enterobacteriaceae. Experiments were performed in triplicate. Data are presented as log10-transformed SSU rDNA gene copy numbers, as interpolated from standard curves of plasmid controls. Error bars represent standard deviations of replicate experiments. NI, non-IBD controls. Control- and IBD-subset are explained in the text. The results of Student's t tests are indicated as *, P < 0.05; **, P < 0.01; and ***, P < 0.001. t tests compared the results of Control- and IBD-subset samples for a particular disease state and bacterial group (e.g., Bacteroidetes/CD/Control subset vs. Bacteroidetes/CD/IBD subset). Data for specific groups were normalized to total bacteria.
Discussion
The chronic inflammation characteristic of UC and CD is suggestive of an inability on the part of patients to control basal level inflammation within gut-associated lymphoid tissues. The antigens responsible for immune system overstimulation are unknown but are likely to arise from the plethora of bacteria that reside within the human GI tract. In this study, we have used culture-independent phylogenetic analysis to characterize the GI microbiotas of IBD patients and non-IBD controls. The surgical samples analyzed provide access to the gut-wall-associated microbiotas of subjects, which may play more critical roles than do fecal microbes in IBD pathogenesis (45, 46). We acknowledge that this census reflects SSU rRNA gene copy number rather than true cell counts, and that the results are subject to biases inherent in PCR amplification and cloning. Furthermore, rRNA analysis will not detect functional changes in GI microbes, such as enhanced virulence, mucosal adherence, or invasion, that do not influence the relative proportions of species in the microbiota (45, 46, 51). Nonetheless, the results provide an internally consistent culture-independent assessment of the gut-wall-associated microbiota.
We report an in-depth molecular survey of microbiology within the human small intestine. Sequence analysis indicates that the wall of the distal small bowel is colonized by microbial populations that are not radically different from those of the large bowel. Both environments are dominated by bacteria of the phyla Bacteroidetes and Firmicutes but differ in the relative proportions in which subgroups of these phyla are present. Members of the class Bacillus (predominantly the Lactobacillales clade) were significantly more abundant in the small intestine than in the colon. In contrast, a variety of members of the Bacteroidetes and the family Lachnospiraceae were less abundant in the small intestine. Moreover, the small-intestine samples, as a whole, contained less overall phylogenetic diversity of Bacteroidetes and Firmicutes than did the large intestine.
The role of MAP in CD pathogenesis continues to be the subject of contentious debate. Of the >15,000 SSU rRNA genes analyzed in this study, only 10 were from Mycobacterium spp., and all were more closely related to species other than MAP. Q-PCR assays of IBD and non-IBD genomic DNAs with MAP-specific (IS900) or M. avium-specific (rRNA) primers were negative (data not shown). Although these data cannot disprove the hypothesis that MAP causes CD, they establish upper limits on the abundance of this potential pathogen in the sample set.
The results of our bacterial survey (i) confirm previous reports of compositional shifts in IBD-associated GI microbial communities and (ii) fail to identify any individual species that is enriched in grossly abnormal IBD samples in a manner suggestive of an active etiologic agent. Recent studies of Manichanh et al. (41) and Gophna et al. (42) also have reported decreased loads of Lachnospiraceae in small sets (approximately six) of CD fecal samples or biopsies in comparison to UC and healthy controls. Our results demonstrate that a distinct subpopulation of IBD patients (the IBD-subset) harbor GI microbiotas that differ significantly from study controls and other CD and UC samples (Control-subset). Neither UC nor CD is characterized by a uniform, stereotypical microbiota. Because the phylogenetic distribution of Control-subset sequences is similar to that observed for healthy adults (32, 44), these samples likely are representative of healthy GI microbiotas. By this criterion, subjects classified in the IBD-subset harbor GI microbiotas that are significantly perturbed in composition relative to “normal” microbiotas.
Because uniform levels of Enterobacteriaceae were measured by Q-PCR in IBD- and Control-subset samples, it is unlikely that contamination of select samples resulted in skewed assessments of microbial diversity. Furthermore, reported patterns of recent (<2 years) antibiotic use, anatomical site of sampling, and gross pathology were not correlated with the microbiotas of IBD- or Control-subset samples. We therefore conclude that the observed differences in the microbiotas of IBD- and Control-subset individuals are genuine and not the result of trivial experimental errors or variations in patient treatment.
The results demonstrate significant perturbations in the GI microbiotas of select IBD patients and thereby suggest that UC and CD are manifest in different forms that can be differentiated by microbial populations. Bacteroidetes and Lachnospiraceae were depleted 1.7 and 2.5 orders of magnitude, respectively, in specimens of the IBD subset. This is noteworthy, because these kinds of organisms promote GI health in myriad ways (30, 52). Several of the OTUs underrepresented in the IBD-subset are most closely related to cultured isolates of butyrate-producing bacteria (Table 1 and SI Table 5). Butyrate and other short-chain fatty acids, which are produced primarily by bacterial metabolism, are important sources of energy for colonic epithelial cells and may enhance epithelial barrier integrity and modulate the GI immune response (53–58). Loss of similar organisms has been correlated with antibiotic-associated diarrhea in a patient (59). We therefore propose a model in which dramatic reductions in the quantities of microbes that provide metabolic services to the host GI tract exacerbate some forms of IBD.
Our data do not address the causal relationships between the reported microbial community imbalances and IBD pathophysiology. Regardless of ultimate causation, the observed microbial imbalances, whether because of prolonged drug treatment or other factors, likely contribute to disease severity. That the hypothesized stratification of UC and CD patients on the basis of the observed microbial imbalances is clinically relevant is suggested by two observations. First, abnormal microbiotas were significantly correlated with the occurrence of abscesses in CD patients. The ability of the commensal microbiota to competitively exclude pathogens may have been disrupted in these cases, thus potentiating abscess formation. Second, patients with abnormal commensal microbiotas were significantly younger than those with normal microbiotas (8.1 years; P = 0.002) at the time of surgery. Loss of the normal GI microbiota might therefore be indicative of more-aggressive forms of IBD. Diet, long-term disease history, and human genetic factors (e.g., CARD15/NOD2 alleles) were not monitored in this study but constitute possible factors underlying the imbalances in selected commensal populations. Although antibiotic treatment per se was not correlated with the observed differences in microbial communities, patients' responses to treatment likely differed; perhaps individuals in the IBD subset were impaired in their capacities to reconstitute normal microbiotas after extensive medical treatment. Remediation of GI microbiotas in these cases, for example through application of specific probiotic regimens, might therefore ameliorate disease (60).
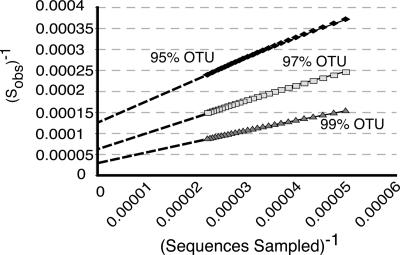
In addition to this study, at least two other large-scale culture-independent studies of the human GI tract have been reported (32, 44). Taken together, the >45,000 bacterial SSU rRNA genes surveyed reveal an unprecedented diversity of microbes associated with the human GI tract. Nearly all of these sequences (>98%) belong to only four bacterial phyla: Firmicutes (64%), Bacteroidetes (23%), Proteobacteria (8%), and Actinobacteria (3%). The lower taxonomic divisions, however, are quite diverse, and collector's curves (SI Fig. 13) do not plateau, indicating that much diversity remains uncharacterized. Nevertheless, we can place lower bounds on the richness of the human GI microbiome both by the Schao1 estimator (61) and by modeling bootstrap resampled collector's curves in a manner analogous to the use of Lineweaver–Burk plots in enzymology (Fig. 5). Both methods estimate that the human gut microbiome consists of at least 1,800 genera and ≈15,000–36,000 species of bacteria, depending on whether species are conservatively (97% OTUs) or liberally (99% OTUs) classified (SI Table 9). Extant sequences represent, at best, 50% of the predicted species-level diversity. Thus, despite the collective depth of these studies, a staggering level of microbial diversity remains to be characterized within the human gut environment.
Fig. 5.
Microbial diversity of the human intestine. OTU richness was estimated for a data set of sequences obtained in this study and those of Eckburg et al. (44) and Ley et al. (32). Sequences were clustered into OTUs and averaged collector's curves simulated by 1,000 random replicate samplings of the OTUs at sample sizes ranging from 1,000 to 45,000 sequences (total sequences, 45,172). The graph shows double reciprocal plots of mean Sobs−1 vs. sequences−1 for 99%, 97%, and 95% OTUs. The Y intercepts of the graphs were extrapolated by linear regression of Sobs−1 and sequences−1, from which expected OTU richness (i.e., Smax) was inferred. The dashed lines extending from each graph approximate the extrapolations.
Materials and Methods
Patients and Sample Collection.
IBD was diagnosed on the basis of combined gross and microscopic features. After surgery at Mt. Sinai School of Medicine, 1.5 cm2 of resected tissue was collected from each patient, placed in tubes containing ethanol/phenol/H2O, and shipped to Boulder, CO. The protocol was approved by the Institutional Review Boards of Mt. Sinai School of Medicine and the University of Colorado. The conditions that defined the negative control group are listed in SI Text.
Library Construction and Sequence Analysis.
Molecular methods used standard protocols (62), elaborated in SI Text. DNA extraction and PCR steps included negative controls for detection of contamination by exogenous DNA. Multiple PCRs were performed for each sample, and products were pooled for further analysis. Species identifications were made by batch BLAST of rRNA databases and more detailed phylogenetic analyses of aligned sequences.
Q-PCR.
Details of Q-PCR are described in SI Text. Plasmid quantification standards were prepared from representative clones of target organisms. Each Q-PCR experiment assayed all DNA templates under consideration and duplicate reactions of the plasmid dilution series and multiple negative-template controls. Quantification of template concentrations was made by linear extrapolation of data from standard curves. Q-PCR experiments were performed in triplicate for each primer set.
Statistical Comparisons of Communities.
Statistical analyses, performed with the R software package (ver. 2.0.1), are described in greater detail in SI Text. For these analyses, sequence data were encoded as presence/absence of OTUs for each sample. Cluster analysis of samples used an agglomerative hierarchical clustering algorithm (63). Associations between medical treatments and membership in the IBD or Control subsets were examined by Fisher's exact test. Q-PCR data were normalized to human actin levels (measured for each sample) and analyzed by two-tailed Student's t test, without treating variances as equivalent. In the cases of specific bacterial groups, Q-PCR results were first normalized to total bacterial loads. The significance of environment clusters was assessed by 1,000 jack-knife resamplings by using the normalized weighted UniFrac test (50).
Supplementary Material
Acknowledgments
We thank Dr. D. Lang (National Institute of Allergy and Infectious Diseases (NIAID)/National Institute of Environmental Health Sciences) for enthusiastic support and Dr. G. Carey, M. Amjadi, T. DeSantis, and M. Hamady for technical support. We thank Drs. L. Baumgartner and V. Visconti for valuable editorial comments. This project has been funded in whole or in part with Federal funds from the NIAID, including under contract no. N01-AI-30055.
Abbreviations
- IBD
inflammatory bowel disease
- CD
Crohn's disease
- UC
ulcerative colitis
- SSU rRNA
small subunit rRNA
- OTU
operational taxonomic unit
- MAP
Mycobacterium avium spp. Paratuberculosis
- PCA
principal components analysis
- Q-PCR
quantitative PCR.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF695452–EF710623).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706625104/DC1.
References
- 1.Farrell RJ, Peppercorn MA. Lancet. 2002;359:331–340. doi: 10.1016/S0140-6736(02)07499-8. [DOI] [PubMed] [Google Scholar]
- 2.Hendrickson BA, Gokhale R, Cho JH. Clin Microbiol Rev. 2002;15:79–94. doi: 10.1128/CMR.15.1.79-94.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanahan F. Lancet. 2002;359:62–69. doi: 10.1016/S0140-6736(02)07284-7. [DOI] [PubMed] [Google Scholar]
- 4.Prantera C, Korelitz BI. Crohn's Disease. New York: Dekker; 1996. [Google Scholar]
- 5.Blumberg RS, Saubermann LJ, Strober W. Curr Opin Gastroenterol. 1999;11:648–656. doi: 10.1016/s0952-7915(99)00032-1. [DOI] [PubMed] [Google Scholar]
- 6.Pizarro TT, Arseneau KO, Cominelli F. Am J Physiol. 2000;278:G665–G669. doi: 10.1152/ajpgi.2000.278.5.G665. [DOI] [PubMed] [Google Scholar]
- 7.Rennick DM, Fort MM. Am J Physiol. 2000;278:G829–G833. doi: 10.1152/ajpgi.2000.278.6.G829. [DOI] [PubMed] [Google Scholar]
- 8.Sartor RB. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 9.Dianda L, Hanby AM, Wright NA, Sebesteny A, Hayday AC, Owen MJ. Am J Pathol. 1997;150:91–97. [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto S, Okabe Y, Setoyama H, Takayama K, Ohtsuka J, Funahashi H, Imaoka A, Okada Y, Umesaki Y. Gut. 1998;43:71–78. doi: 10.1136/gut.43.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onderdonk AB, Franklin ML, Cisneros RL. Infect Immun. 1981;32:225–231. doi: 10.1128/iai.32.1.225-231.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE, Balish E, Taurog JD, Hammer RE, Wilson KH, Sartor RB. J Clin Invest. 1996;98:945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, Balish E, Hammer RE. J Exp Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper PH, Lee EC, Kettlewell MG, Bennett MK, Jewell DP. Gut. 1985;26:279–284. doi: 10.1136/gut.26.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberhelman HA, Jr, Kohatsu S, Taylor KB, Kivel RM. Am J Surg. 1968;115:231–240. doi: 10.1016/0002-9610(68)90034-2. [DOI] [PubMed] [Google Scholar]
- 17.Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, Kerremans R, Vantrappen G. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 18.Winslet MC, Allan A, Poxon V, Youngs D, Keighley MR. Gut. 1994;35:236–242. doi: 10.1136/gut.35.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prantera C, Scribano ML. Ital J Gastroenterol Hepatol. 1999;31:244–246. [PubMed] [Google Scholar]
- 20.Van Kruiningen HJ. Inflamm Bowel Dis. 1999;5:183–191. doi: 10.1097/00054725-199908000-00007. [DOI] [PubMed] [Google Scholar]
- 21.El-Zaatari FA, Osato MS, Graham DY. Trends Mol Med. 2001;7:247–252. doi: 10.1016/s1471-4914(01)01983-9. [DOI] [PubMed] [Google Scholar]
- 22.Hermon-Taylor J. Gut. 2001;49:755–756. doi: 10.1136/gut.49.6.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenstein RJ. Lancet Infect Dis. 2003;3:507–514. doi: 10.1016/s1473-3099(03)00724-2. [DOI] [PubMed] [Google Scholar]
- 24.Sartor RB. Gut. 2005;54:896–898. doi: 10.1136/gut.2004.055889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Kelly D, Conway S, Aminov R. Trends Immunol. 2005;26:326–333. doi: 10.1016/j.it.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Bry L, Falk PG, Midtvedt T, Gordon JI. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 29.Stappenbeck TS, Hooper LV, Gordon JI. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooper LV, Midtvedt T, Gordon JI. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 31.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 34.Giaffer MH, Holdsworth CD, Duerden BI. J Med Microbiol. 1991;35:238–241. doi: 10.1099/00222615-35-4-238. [DOI] [PubMed] [Google Scholar]
- 35.Krook A, Lindstrom B, Kjellander J, Jarnerot G, Bodin L. J Clin Pathol. 1981;34:645–650. doi: 10.1136/jcp.34.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merwe JPvd, Schroder AM, Wensick F, Hazenberg MP. Scand J Gastroenterol. 1988;23:1125–1131. doi: 10.3109/00365528809090179. [DOI] [PubMed] [Google Scholar]
- 37.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Dore J. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangin I, Bonnet R, Seksik P, Rigottier-Gois L, Sutren M, Bouhnik Y, Neut C, Collins MD, Colombel J-F, Marteau P, Dore J. FEMS Micro Ecol. 2004;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Prindiville T, Cantrell M, Wilson KH. Inflamm Bowel Dis. 2004;10:824–833. doi: 10.1097/00054725-200411000-00017. [DOI] [PubMed] [Google Scholar]
- 40.Lepage P, Seksik P, Sutren M, de la Cochetiere MF, Jian R, Marteau P, Dore J. Inflamm Bowel Dis. 2005;11:473–480. doi: 10.1097/01.mib.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- 41.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 46.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenstein RJ, Greenstein AJ. Mol Med Today. 1995;1:343–348. doi: 10.1016/s1357-4310(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 48.Mishina D, Katsel P, Brown ST, Gilberts EC, Greenstein RJ. Proc Natl Acad Sci USA. 1996;93:9816–9820. doi: 10.1073/pnas.93.18.9816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lozupone C, Knight R. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 52.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 53.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 54.Galvez J, Rodriguez-Cabezas ME, Zarzuelo A. Mol Nutr Food Res. 2005;49:601–608. doi: 10.1002/mnfr.200500013. [DOI] [PubMed] [Google Scholar]
- 55.Scheppach W, Weiler F. Curr Opin Clin Nutr Metab Care. 2004;7:563–567. doi: 10.1097/00075197-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 56.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 57.Lara-Villoslada F, de Haro O, Camuesco D, Comalada M, Velasco J, Zarzuelo A, Xaus J, Galvez J. Eur J Nutr. 2006;45:418–425. doi: 10.1007/s00394-006-0610-2. [DOI] [PubMed] [Google Scholar]
- 58.Segain JP, Raingeard de la Bletiere D, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottiere HM, Galmiche JP. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young VB, Schmidt TM. J Clin Microbiol. 2004;42:1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sartor RB. Gastroenterology. 2004;126:1620–1633. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 61.Chao A. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 62.Dalby AB, Frank DN, St Amand AL, Bendele AM, Pace NR. Appl Environ Microbiol. 2006;72:6707–6715. doi: 10.1128/AEM.00378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.