Abstract
Extensive neurogenetic analysis has shown that memory formation depends critically on cAMP-protein kinase A (PKA) signaling. Details of how this pathway is involved in memory formation, however, remain to be fully elucidated. From a large-scale behavioral screen in Drosophila, we identified the yu mutant to be defective in one-day memory after spaced training. The yu mutation disrupts a gene encoding an A-kinase anchoring protein (AKAP). AKAPs comprise a family of proteins, which determine the subcellular localization of PKAs and thereby critically restrict cAMP signaling within a cell. Further behavioral characterizations revealed that long-term memory (LTM) was disrupted specifically in the yu mutant, whereas learning, short-term memory and anesthesia-resistant memory all appeared normal. Another independently isolated mutation of the yu gene failed to complement the LTM defect associated with the yu mutation, and this phenotypic defect could be rescued by induced acute expression of a yu+ transgene, suggesting that yu functions physiologically during memory formation. AKAP Yu is expressed preferentially in the mushroom body (MB) neuroanatomical structure, and expression of a yu+ transgene to the MB, but not to other brain regions, is sufficient to rescue the LTM defect of the yu mutant. These observations lead us to conclude that proper localization of PKA by Yu AKAP in MB neurons is required for the formation of LTM.
Keywords: A-kinase anchoring protein, cAMP pathway, mushroom body, olfactory memory
Extensive studies have shown that memory formation depends critically on the cAMP-protein kinase A (PKA) signaling pathway in vertebrates and invertebrates (1, 2). The subcellular location of PKA is determined by a family of A-kinase anchoring proteins (AKAPs), which are defined by their ability to copurify with PKA activity (3). These molecules are thought to serve as signal-organizing molecules and play a critical role in specifying functions of the cAMP pathway by compartmentalizing PKA with other signaling molecules. AKAPs have been reported to be involved in synaptic plasticity by the interaction with the regulatory subunits of PKA (4). For instance, in Aplysia, disruption of an AKAP::PKA-RII interaction by the inhibitory peptide, Ht31, prevents both the short- and long-term facilitation induced by serotonin (5-HT) (5). In rat, infusion into the lateral amygdale of inhibitory peptides that compete with PKA regulatory subunits for binding with AKAPs affects auditory fear memory (6). To date, however, no specific AKAP has been shown to be involved in memory formation.
Memory in Drosophila typically has been studied with a Pavlovian olfactory task in which an odor is temporally associated with foot-shock punishment (7) and has been extensively characterized at the molecular and anatomical levels (2, 8–10). Genetic dissection of the conditioned behavioral response has suggested several temporal phases, including acquisition (or learning, LRN), short-term memory (STM), middle-term memory (MTM), anesthesia-resistant memory (ARM), and protein synthesis-dependent long-term memory (LTM) (11). STM and MTM appear labile and short-lived, whereas ARM and LTM persist for several days. LTM is disrupted, but STM, MTM, and ARM are normal, in flies fed the protein synthesis inhibitor, cycloheximide (CXM). One training session and ten massed training (no intersession rest interval) produce only a decremental and CXM-insensitive memory (STM, MTM, and ARM), but 10 spaced training sessions (a 15-min inter-session rest interval) produces STM, MTM, ARM and a protein synthesis-dependent LTM that persists for more than a week. We use one-day memory to determine ARM and LTM for massed training one-day memory contains only ARM, whereas for spaced training, one-day memory consists of ARM and LTM. STM and MTM have diminished 1 d after either massed or spaced training (11).
The cAMP signaling pathway is important for memory formation. The STM mutants, rutabaga (rut) (12) and dunce (dnc) (13), encodes adenylyl cyclase (AC) and cAMP-specific phosphodiesterase (PDE), respectively. Mutation of the PKA-RI gene disrupts both learning and memory (14). A temperature-sensitive mutant of PKA-C1 (DCO) disrupts MTM (15). And, disruption of the transcription factor CREB (cAMP responsive element binding protein) specifically blocks LTM (16). Some of these memory-related functions of cAMP signaling have been mapped to the mushroom body (MB). Expression of a rut+ transgene in the MB is sufficient to rescue the STM defects of the rut mutant (17), and overexpression of CREB repressor in the MB blocks LTM formation (18).
From a large-scale behavioral screen for mutants with defects in one-day memory after spaced training, we have identified the yu mutant (yu means “foolish” in Chinese). LRN, STM, MTM, and ARM are normal, but LTM specifically is defective, in the yu mutant. The yu mutation disrupts a gene encoding an AKAP. Using anti-Yu antibody, we show that this gene is expressed preferentially in the MB. With a yu+ transgene, we also show that expression of Yu in the MB is sufficient to rescue the LTM defect of the yu mutant.
Results
Disruption of yu Specifically Impairs LTM.
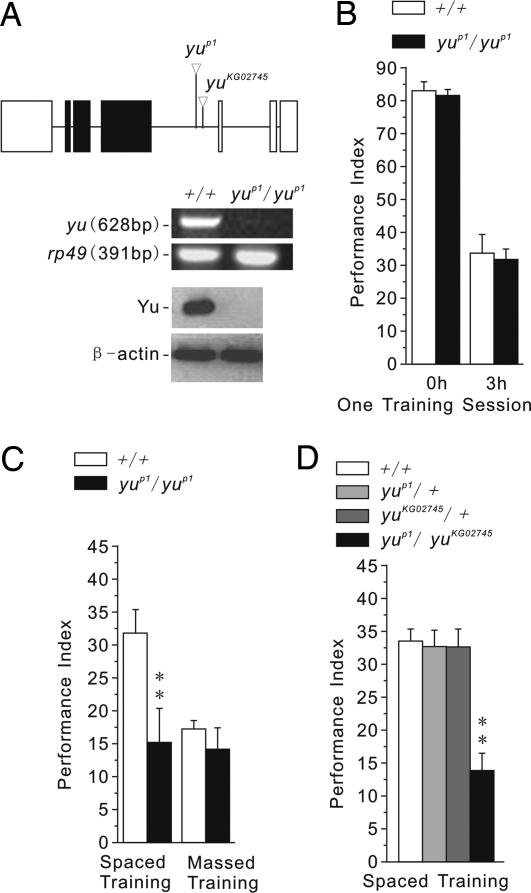
We conducted a P-element mutagenesis to screen behaviorally for X-linked mutants with defective one-day memory after spaced training. Eleven mutants were identified from 2,021 homozygous-viable transposant lines. By means of plasmid rescue and DNA sequencing, we determined the mutant, yuP1, to carry a P-element insertion in an intron of the CG3249 gene, which encodes a putative AKAP (Fig. 1A Top) (19). RT-PCR and Western blot analysis revealed that yu transcript and protein were undetectable in yuP1 mutants (Fig. 1A Middle and Bottom).
Fig. 1.
LTM is specifically impaired in yu mutants. (A) Molecular-genetic characterization of yuP1 mutant. (Top) Plasmid rescue and sequencing of genomic DNA flanking the P-element insertion site indicated the P-element was inserted in the intron of CG3249, which encodes a novel putative AKAP. Perusal of the Bloomington Stock Center identified a second P-element insertion, yuKG02745, near the insertion site of yuP1. Blank box represents an untranslated region, the black box represents a protein coding region, and the line represents an intron. The transcription direction is from right to left. (Middle) RT-PCR experiments showed that the yu transcript was greatly reduced in yuP1/yuP1 mutants. Ribosomal protein 49 (rp49) was used as a loading control. (Bottom) Western blot analysis showed that Yu protein also was reduced greatly in yuP1/yuP1 mutants. β-Actin was used as a loading control. Left blots are control flies (+/+); right blots are yuP1/yuP1 mutants. (B) Memory retention, measured immediately or 3 h after one training session, was normal in yuP1/yuP1 mutants (n = 8 PIs per group). All PIs of these and the following behavioral experiments are means and SEM. Statistical significances are determined from t test. Asterisks indicates critical values of *, P < 0.05 and **, P < 0.01. (C) One-day memory after spaced training was significantly impaired, but that after massed training was normal, in yuP1/yuP1 mutants (n = 8 PIs per group). (D) Genetic complementation analysis between yuP1 and yuKG02745 for one-day memory after spaced training revealed the two mutations to be recessive to wild-type (+) and allelic (noncomplementing). Thus, the yu gene was involved in LTM formation (n = 8 PIs per group).
Memory retention in mutant yuP1 flies was normal when measured immediately or 3 h after one training session (Fig. 1B). Similarly, one-day memory after massed training (ARM) was normal, but one-day memory after spaced training (ARM and LTM) was impaired (Fig. 1C). One-day memory after spaced training, in fact, was similar to that after massed training in the yu mutant. Sensorimotor responses to the odors and foot-shock used for the Pavlovian experiments also were normal in yu mutants (Table 1). Together, these results indicate that STM, MTM, and ARM are normal, but LTM specifically is impaired, in the yuP1 mutant.
Table 1.
Sensorimotor responses of different genotypes to the odors and foot-shock used for Pavlovian learning
| Genotype | OA (OCT) | OA (MCH) | SR |
|---|---|---|---|
| W1118 (isoCJ1) | 76 ± 1.4 | 76 ± 2.7 | 83 ± 2.1 |
| W1118 (isoCJ1) (+HS) | 71 ± 2.3 | 75 ± 1.2 | 83 ± 2.3 |
| yuP1/yuP1 | 77 ± 2.9 | 76 ± 2.4 | 80 ± 1.3 |
| yuKG02745/yuKG02745 | 74 ± 3.1 | 78 ± 2.3 | 82 ± 1.9 |
| yuP1/yuP1; hs-yu+/hs-yu+ | 71 ± 3.3 | 74 ± 2.6 | 84 ± 2.6 |
| yuP1/yuP1; hs-yu+/hs-yu+ (+HS) | 73 ± 1.0 | 73 ± 2.9 | 81 ± 0.7 |
No significant differences in ″task-relevant″ sensorimotor responses were detected for comparisons between control flies and mutants or other experimental genotypes (in each case, n = 8 for all the genotypes). All experiments were conducted at RT. OA, olfactory acuity; OCT, octanol; MCH, methylcyclohexanol; SR, shock reactivity; HS, a 30-min heat shock (37°C), followed by a 3-h recovery period.
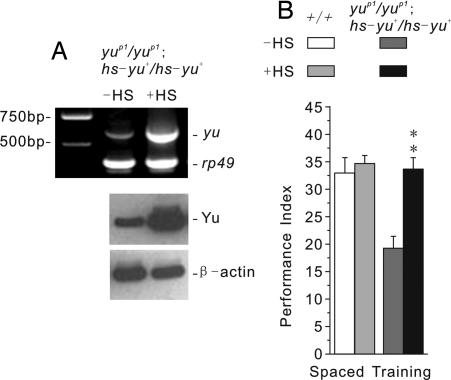
To confirm that this memory defect results from disruption of CG3249, we conducted a genetic complementation experiment using yuP1 and another existing P-element insertion allele, yuKG02745 (Fig. 1A Top) (19). Although yuP1/+ and yuKG02745/+ heterozygotes showed normal one-day memory after spaced training, such memory was defective in yuP1/yuKG02745 heteroallelic flies (Fig. 1D). We also constructed transgenic yuP1 mutants carrying an inducible hs-yu+ construct. Acute expression of this transgene 3 h before training was sufficient to induce high levels of yu transcript and protein (Fig. 2A) and to rescue the memory defect in yuP1 mutants (Fig. 2B). These latter observations demonstrate that the memory defect of the yu mutant does not result from abnormal development. Instead, Yu appears to be involved physiologically in the adult brain during the formation of LTM.
Fig. 2.
Acute expression of yu+ transgene in the mutant background can rescue the LTM defect. (A) mRNA and protein expression levels of yu+ in yuP1/yuP1; hs-yu+/hs-yu+ transgenic mutants immediately or 3 h after a 30-min heat shock treatment (37°C), respectively. (Upper) RT-PCR revealed strong overexpression of yu+ after a heat shock treatment. Left column, DNA makers; center column, transgenic mutants with no heat shock treatment (−HS); right column, transgenic mutants after heat shock treatment (+HS). Rp49 was used as a loading control. (Lower) Western blot analysis showed strong overexpression of Yu after heat shock treatment. Left column, transgenic mutants with no heat shock treatment (−HS); right column, transgenic mutants after heat shock treatment (+HS). β-actin was used as a loading control. (B) The defect in one-day memory after spaced training was rescued in yuP1/yuP1; hs-yu+/hs-yu+ transgenic mutants when animals were trained 3 h after a 30-min heat shock treatment (37°C) (n = 8 PIs per group).
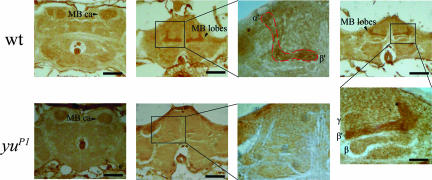
Yu is expressed preferentially in the MB. To gain insights into the neural basis for this genetic dissection of LTM formation by Yu AKAP, we performed immunohistochemistry analysis. Yu appears preferentially expressed in the MB, α′ and β′ lobes in particular (Fig. 3), and the staining in the rest of the brain is minimal and similar that seen for the mutant where Western blot analysis suggests no expression (Fig. 1A Bottom). These observations suggested the hypothesis that Yu functions in the MB during the formation of LTM.
Fig. 3.
Yu is expressed preferentially in the MB. (Upper) Frontal sections of wild-type adult fly brains after immunostaining (dark brown) with an affinity-purified antiserum against a peptide fragment of Yu. Mild signal was detected in sections through the MB calyx regions (CA). Stronger signal was apparent in the MB, especially in regions of α′/β′ lobes (MB). (Lower) Frontal sections of yuP1 mutant adults after immunostaining with the Yu antibody. Yu expression in the MB was greatly reduced. (Scale bar, 50 μm.)
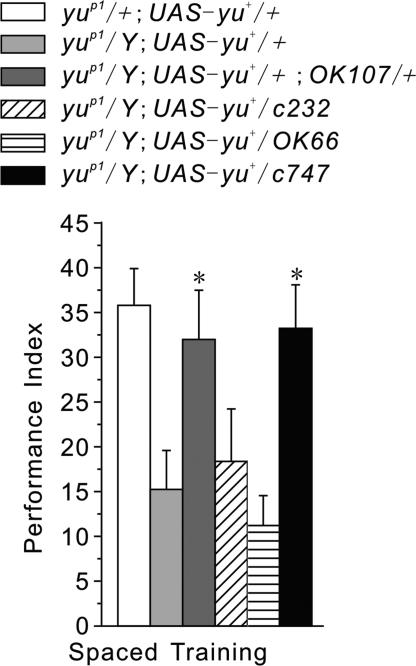
Expression of yu+ in the MB rescues the LTM defect of yu mutants. To map Yu-related functional brain regions, we used several Gal4 lines to target expression of UAS-yu+ in specific neuroanatomical regions associated with memory (20). OK107-Gal4 (OK107) and c747-Gal4 (c747) is highly specific to MB with strong expression in a majority of MB neurons (21, 22), whereas OK66-Gal4 (OK66) is mainly expressed in antennal lobe (AL) local neurons with weak expression in a small population of MB neurons (23), and c232 shows rather exquisite expression restricted to the central complex (24). OK107- or c747-driven expression of UAS-yu+ in the MB of the yuP1 mutant was sufficient to restore normal LTM (Fig. 4). In contrast, the LTM defect of the yuP1 mutant remained when UAS-yu+ transgenic expression was driven by OK66 in antennal lobe or by c232 in central complex (Fig. 4). These observations lead us to conclude that the function of Yu AKAP in the MB is essential for the formation of LTM.
Fig. 4.
LTM formation depends on yu expression in the MB. One-day memory after spaced training was evaluated in transgenic yuP1 mutants with various Gal4 drivers expressing a UAS-yu+ transgene. The LTM defect of yu mutants was rescued when UAS-yu+ was expressed in the MB (yuP1/Y; UAS-yu+/+; OK107/+ and yuP1/Y; UAS-yu+/+; c747/+). The LTM defect was not rescued, however, when UAS-yu+ was expressed in the AL (yuP1/Y; UAS-yu+/+; OK66/+) or in CC (yuP1/Y; UAS-yu+/+; c232/+). Transgenic yuP1/+; UAS-yu+/+ flies were controls for genetic background and leaky UAS-driven transgene effects (n = 8 PIs per group).
Yu in the MB Interacts with cAMP-Dependent Signaling During the Formation of LTM.
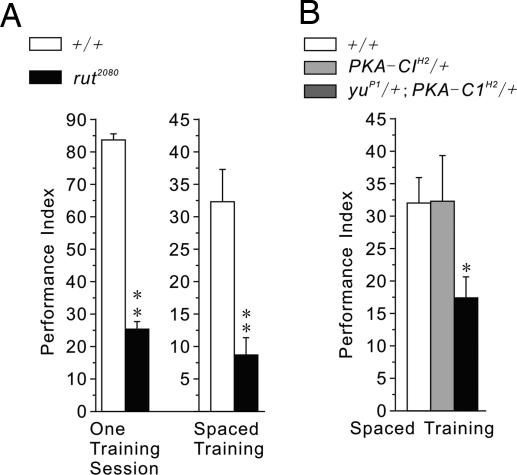
Expression of Rut in the MB is known to be critical for STM (17), whereas PKA-C1 is expressed highly in the MB and its mutations thereof have been shown to affect both learning and MTM (25). We show here that one-day memory after spaced training is also disrupted in rut mutants (Fig. 5A). More pertinently, we also have established an epistatic interaction between Yu and PKA-C1. Flies heterozygous for either gene (PKA-C1H2/+ or yuP1/+) displayed normal one-day memory after spaced training, but such memory was impaired in the double-heterozygous flies (yuP1/+; PKA-C1H2/+) (Figs. 1D and 5B). This genetic observation implies that Yu and PKA-C1 function together in the same cAMP pathway during the formation of LTM.
Fig. 5.
Yu-mediated cAMP signaling in the MB contributes to the formation of LTM. (A) Both immediate memory after one training session and one-day memory after spaced training were impaired in rut2080 mutants (n = 6 or 8 PIs per group for experiments using one training session or spaced training, respectively). (B) One-day memory after spaced training was normal in heterozygotes carrying mutations of PKA (PKA-C1H2/+) or of yu AKAP (yuP1/+) but significantly disrupted in double heterozygotes (yuP1/+; PKA-C1H2/+) (n = 6 PIs per group).
Discussion
Studies in several species have revealed a time-dependent process of memory consolidation with distinct temporal phases of memory (26–28). After Drosophila olfactory learning, behavioral, pharmacological, and genetic manipulations have “dissected” memory formation into four distinct but interdependent phases: STM, MTM, ARM, and LTM (11). Our current study focuses on the role of a specific AKAP gene, yu, in formation of LTM. We have shown that (i) LTM specifically was abolished in yu mutants, whereas other memory phases appeared normal (Fig. 1 B and C), (ii) this LTM defect was produced by (independent) mutations in the yu transcription unit, CG3249 (Fig. 1D), and (iii) the LTM defect of the yu mutant could be rescued via acute induction of a yu+ transgene (Fig. 2B). These data clearly define a specific, physiological function for Yu AKAP during memory formation.
Immunostaining of adult brain has revealed that Yu is expressed preferentially in the MB anatomical region (Fig. 3), which many studies have established plays a key role in olfactory memory formation (2, 8, 29, 30). Using multiple “enhancer trap” GAL4-drivers, with overlapping patterns of expression in MB or with preferential expression in other potentially critical anatomical regions [i.e., antennal lobes (31, 32) and central complex (33)], we have spatially restricted the expression of a UAS-yu+ transgene in the yu mutant. LTM was normal only when Yu was expressed in the MB (Fig. 4). Finally, a classically designed genetic epistasis experiment has shown that Yu AKAP interacts with PKA-C1 during LTM formation (Fig. 5B). This synergistic effect supports the idea that Yu functions as an AKAP in mediating LTM formation.
Although genetic dissection has revealed distinct memory phases and gene disruption experiments have identified several components of cAMP signaling to be important for this process, few studies have determined whether these molecules actually function together during memory formation. Rut-AC and PKA-C1, for instance, are critical not only for learning and STM or MTM (12, 15, 25, 34) but also for LTM (Fig. 5 A and B). But, do they function together in the same place during each memory phase? When considering AKAPs, more specifically, proteins of this family bind regulatory subunits of PKA, and thereby may serve to localize PKA to different subcellular compartments (35). But specifically which AKAP is involved in which cellular function in Drosophila (19, 36–38)? Our work on the Yu AKAP presents the hypothesis that localization of Yu helps to define a function of cAMP signaling specific to LTM formation. As a corollary, we speculate that other AKAPs (in the MB) may introduce specificities for cAMP signaling during earlier memory phases by means of differential localization of PKA to other subcellular compartments.
Although both STM and LTM are localized in the MB, these two memory components may not reside in the same population of MB neurons. There are evidences that the horizontal lobe is especially important for Rut-dependent STM formation (24, 39), whereas vertical (α and α′) lobes are critical for LTM formation (40). Previous studies have shown that Drosophila α/β neurons not only function in the memory retrieval (41, 42) but also form a branch-specific LTM trace after spaced training (18). Functional analysis of subsets of MB neurons show that α′/β′ neurons are required to acquire and stabilized an olfactory memory (42). To that end, our study also shows that Yu is expressed preferentially in α′ and β′ lobes.
Methods
Isolation of Memory-Defective Mutants.
Transposon mutagenesis was carried out as described in previous works with minor modifications (43). A PlacW transposon, carried on the second chromosome, was used as a mutator to generate homo- or hemizygous-viable X-linked transposants [see supporting information (SI) Fig. 6 and SI Methods]. One-day memory after spaced training was quantified (n = 1) for 2,021 transposant strains. Two hundred fifty-nine mutants produced PIs that were <50% of the mean PI for w1118 (isoCJ1) control flies. PIs (n = 4) then were generated for these strains, and 63 of them yielded mean PIs <50% of the mean PI of control flies. The 63 low-scoring mutants were back-crossed for five generations to control flies to equilibrate genetic backgrounds. Out-crossed strains again were tested for one-day memory after spaced training and for olfactory acuity and shock reactivity. The yuP1 mutant was one such strain that retained poor one-day memory after spaced training and showed normal olfactory acuity and shock reactivity.
Drosophila Strains and Genetics.
The KG02745 (19) were obtained from the Bloomington Stock Center (Bloomington, IN). Mutant rut2080 (33) flies were obtained from the laboratory of Li Liu at the Institute of Biophysics CAS (Beijing, China). OK107 (MB-Gal4), c747 (MB-Gal4), c232 (central complex Gal4, CC-Gal4) were extant stocks (out-crossed) in our laboratory. To equilibrate genetics backgrounds, these stocks KG02745 were out-crossed with “cantonized” FM7 flies carrying an X-chromosome and autosomes from the w1118 (isoCJ1) control flies. All other stocks including OK66, hs-yu+, and UAS-yu+ were equilibrated by five generations of out-cross to w1118 (isoCJ1).
Plasmid Rescue of P-Element Insertion.
DNA sequences flanking the yuP1 insertion site were identified by digesting total genomic DNA from yuP1 mutants with either EcoRI or XbaI, ligating the resulting restriction fragments and then transforming bacteria to isolate ampicillin-resistant plasmids. Primers corresponding to P-element sequence then were used to direct DNA sequencing into flanking genomic DNA. The insertion site was confirmed by generating PCR primers on either side of the insertion, amplifying the intervening DNA and determining the size of the amplicon (43).
Generation of Transgenic Drosophila Lines.
To create hs-yu+ and UAS-yu+ construct, the expected full-length yu+ cDNA was amplified by Pfu-polymerase from cDNA plasmid GM14014 (from Drosophila Genomics Resource Center, DGRC). The amplified fragment (≈3.3 kb) was subcloned into pGEM-T (Promega, Madison, WI) vector and sequenced to confirm the integrity of the amplified cDNA fragment. The subcloned yu+ fragment then was released from pGEM-T vector by restriction digest with NotI (NEB, Beverly, MA). The gel-purified fragment finally was subcloned into either a NotI-cut and calf intestinal alkaline phosphatase (CIP; NEB)-treated CaSpeR-hs (8.9 kb) or a pUAST (9 kb) germ-line transformation vectors. The direction of subcloning and sequence were confirmed. Germ-line transformation was carried out as described by Rubin and Spradling (44).
Pavlovian Olfactory Learning.
The training and testing procedures were the same as described (7, 11, 16). Briefly, a group of 100 flies was sequentially exposed for 60 s to each of two odors (octanol (OCT) or methylcyclohexanol (MCH), with 45 seconds of fresh air in between). Flies were subjected to foot-shock (12 1.5-s pulses with 3.5-s intervals, 60 V) during exposure to the first odor (CS+: OCT) but not to the second (CS−; MCH). To measure “immediate memory,“ flies were transferred immediately after training to the choice point of a T-maze and forced to choose between the two odors for 2 min, at which time they were trapped in their respective T-maze arms, anesthetized, and counted. A reciprocal group of flies was trained and tested by using MCH as the CS+ and OCT as the CS−. PIs from these two groups finally were average for an n = 1. A PI of 0 represented a 50:50 distribution, whereas a PI of 100 represented 100% avoidance of the shock-paired odor (7). Spaced training consisted of 10 training sessions with a 15-min rest between each. Massed training consisted of 10 training sessions with no rest interval. To quantify one-day memory, flies were transferred after training to food vials and stored at 18°C for 24 h. At least 30 min before being tested, flies were returned to room temperature (RT).
Sensorimotor Responses.
Odor-avoidance responses were quantified by exposing naïve flies to one odor (OCT or MCH) versus air in the T-maze (7). After 2 min, flies were trapped in their respective T-maze arms, anesthetized, and counted. A PI was calculated for each odor individually as reported (7). The ability to sense and escape from foot-shock (shock reactivity) was quantified in naïve flies by inserting electrifiable grids into both arms of the T-maze but delivering shock pulses only in one arm of the T-maze and allowing flies to choose between the two arms. After one minute, flies were trapped in their respective arms, anesthetized, and counted. Individual PIs were calculated as for olfactory acuity.
Heat Shock Protocol.
Flies subjected to heat shock were placed in empty vials in an air incubator that was maintained at 37°C for 30 min (45). Flies then were transferred back to bottles with food and allowed to rest for 3 h at RT (20–24°C) before training. To minimize leaky expression of the transgene for groups not subjected to heat shock, flies were incubated at 18°C overnight before training. All training and testing was performed at RT.
RT-PCR of Heat Shock Induced yu Expression.
Flies were raised at either 18 or 25°C. One group of flies was shifted from 18 to 37°C for 30 min and immediately frozen at −80°C. Total RNA was prepared from 50 frozen flies by using the classic TRIzol method. Three independent RNA preparations were made for each group of flies. First-strand cDNA was synthesized directly from the mRNA by using the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA). PCR amplification with TaqDNA polymerase (Invitrogen) was carried out in Invitrogen 10× PCR buffer by using 1.5 mM MgCl2 with 30 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, followed by extension at 72°C for 10 min. A pair of yu-specific primers were designed against regions of the Drosophila yu cDNA (GenBank accession no. NM_131993). Primers yus1 (5′-aatctagatccagagcaatggacgca-3′) and yus2 (5′-aatctagaacgctggccagggtcttg-3′) amplify a 628-bp fragment. Lack of genomic DNA contamination was confirmed by the absence of any PCR products when RT-PCR was carried out in control reactions without reverse transcriptase. Ribosomal protein 49 (rp49) primers (5′-atgaccatccgcccagcatac-3′ and 5′-gagaacgcaggcgaccgttgg-3′) were designed to amplify a 391-bp fragment between bases 1 and 391 of the rp49-coding region (GenBank accession no. Y13939). The control rp49 mRNA is expressed at equal levels in all cells at all stages (46).
Generation and Purification of Antisera and Western Blot Analysis.
For generation of antisera, rabbits were injected with a purified polypeptide fragment containing Drosophila Yu peptide fragments (TNYTNKECEQNNNCEPKEEP). Anti-Yu antisera were affinity-purified by using Protein A spin chromatography kit (Pierce, Rockford, IL). For Western blot analyses, whole adult flies were frozen in liquid nitrogen and shaken through a sieve to separate the heads. The frozen heads (≈1 ml) were crushed with a mortar and pestle and homogenized with an automatic homogenizer in 5 mM Hepes, 100 mM NaCl (pH 7.4) with protease inhibitors (Complete protease inhibitor mixture; Roche, Indianapolis, IN). Large cuticular debris was removed by centrifugation at 5,000 × g for 20 min at 4°C. Fly extracts (80 μg of protein) were electrophoresed on SDS/polyacrylamide gels and blotted onto NC membranes (Millipore, Bedford, MA). Blots were incubated with affinity-purified anti-Yu sera overnight at 4°C, HRP-conjugated goat-anti-rabbit IgG (Sigma, St. Louis, MO) for 1 h at RT and visualized with SuperSignal Chemiluminscent substrate (Pierce).
Immunohistochemistry.
Immunohistochemistry on adult Drosophila heads was performed on paraffin sections. Details were available in SI Methods.
Supplementary Material
Acknowledgments
We thank the Bloomington Stock Center for providing Drosophila stocks. This work was supported by the National Basic Research Project (973 program) of the Ministry of Science and Technology of China 2006CB500800 (to Y.Z.), National Nature Sciences Foundation of China Grants 30670460, 30429001, and 30221003 (to H.-M.Z.), the Tsinghua-Yue-Yuen Medical Sciences Fund (to Y.Z.), Major International (Regional) Joint Research Project of National Natural Sciences Foundation of China Grant 30220120692 (to Z.X.) and the DART Neuroscience Fund (to T.T and Y.Z.).
Abbreviations
- AKAP
A-kinase anchoring protein
- ARM
anesthesia-resistant memory
- LTM
long-term memory
- MB
mushroom body
- PI
performance index
- PKA
protein kinase A
- MTM
middle-term memory
- RT
room temperature
- STM
short-term memory.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700439104/DC1.
References
- 1.Kandel ER. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 2.Davis RL. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 3.Wong W, Scott JD. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 4.Bauman AL, Goehring AS, Scott JD. Neuropharmacology. 2004;46:299–310. doi: 10.1016/j.neuropharm.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Hu JY, Schacher S, Schwartz JH. J Neurosci. 2004;24:2465–2474. doi: 10.1523/JNEUROSCI.4331-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moita MA, Lamprecht R, Nader K, LeDoux JE. Nat Neurosci. 2002;5:837–838. doi: 10.1038/nn901. [DOI] [PubMed] [Google Scholar]
- 7.Tully T, Quinn WG. J Comp Physiol A. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- 8.Heisenberg M. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 9.Waddell S, Quinn WG. Annu Rev Neurosci. 2001;24:1283–1309. doi: 10.1146/annurev.neuro.24.1.1283. [DOI] [PubMed] [Google Scholar]
- 10.Dubnau J, Tully T. Curr Biol. 2001;11:R240–243. doi: 10.1016/s0960-9822(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 11.Tully T, Preat T, Boynton SC, Del Vecchio M. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 12.Livingstone MS, Sziber PP, Quinn WG. Cell. 1984;37:205–215. doi: 10.1016/0092-8674(84)90316-7. [DOI] [PubMed] [Google Scholar]
- 13.Byers D, Davis RL, Kiger JA., Jr Nature. 1981;289:79–81. doi: 10.1038/289079a0. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin SF, Del Vecchio M, Velinzon K, Hogel C, Russell SR, Tully T, Kaiser K. J Neurosci. 1997;17:8817–8827. doi: 10.1523/JNEUROSCI.17-22-08817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Tully T, Kalderon D. Learn Mem. 1996;2:320–333. doi: 10.1101/lm.2.6.320. [DOI] [PubMed] [Google Scholar]
- 16.Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 17.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 18.Yu D, Akalal DB, Davis RL. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutsuddi M, Marshall CM, Benzow KA, Koob MD, Rebay I. Curr Biol. 2004;14:302–308. doi: 10.1016/j.cub.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Lee T, Lee A, Luo L. Development (Cambridge, UK) 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 22.Dubnau J, Grady L, Kitamoto T, Tully T. Nature. 2001;411:476–480. doi: 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Chiang AS, Xia S, Kitamoto T, Tully T, Zhong Y. Curr Biol. 2003;13:1900–1904. doi: 10.1016/j.cub.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Zars T, Fischer M, Schulz R, Heisenberg M. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 25.Skoulakis EM, Kalderon D, Davis RL. Neuron. 1993;11:197–208. doi: 10.1016/0896-6273(93)90178-t. [DOI] [PubMed] [Google Scholar]
- 26.Kandel ER. Biosci Rep. 2001;21:565–611. doi: 10.1023/a:1014775008533. [DOI] [PubMed] [Google Scholar]
- 27.DeZazzo J, Tully T. Trends Neurosci. 1995;18:212–218. doi: 10.1016/0166-2236(95)93905-d. [DOI] [PubMed] [Google Scholar]
- 28.Abel T, Lattal KM. Curr Opin Neurobiol. 2001;11:180–187. doi: 10.1016/s0959-4388(00)00194-x. [DOI] [PubMed] [Google Scholar]
- 29.Quinn WG. Nature. 2006;439:546–548. doi: 10.1038/439546a. [DOI] [PubMed] [Google Scholar]
- 30.Dubnau J, Chiang AS, Tully T. J Neurobiol. 2003;54:238–253. doi: 10.1002/neu.10170. [DOI] [PubMed] [Google Scholar]
- 31.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 32.Yu D, Ponomarev A, Davis RL. Neuron. 2004;42:437–449. doi: 10.1016/s0896-6273(04)00217-x. [DOI] [PubMed] [Google Scholar]
- 33.Liu G, Seiler H, Wen A, Zars T, Ito K, Wolf R, Heisenberg M, Liu L. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- 34.Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, Reed RR. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- 35.Langeberg LK, Scott JD. J Cell Sci. 2005;118:3217–3220. doi: 10.1242/jcs.02416. [DOI] [PubMed] [Google Scholar]
- 36.Han JD, Baker NE, Rubin CS. J Biol Chem. 1997;272:26611–26619. doi: 10.1074/jbc.272.42.26611. [DOI] [PubMed] [Google Scholar]
- 37.Jackson SM, Berg CA. Development (Cambridge, UK) 2002;129:4423–4433. doi: 10.1242/dev.129.19.4423. [DOI] [PubMed] [Google Scholar]
- 38.Terman JR, Kolodkin AL. Science. 2004;303:1204–1207. doi: 10.1126/science.1092121. [DOI] [PubMed] [Google Scholar]
- 39.Akalal DB, Wilson CF, Zong L, Tanaka NK, Ito K, Davis RL. Learn Mem. 2006;13:659–668. doi: 10.1101/lm.221206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pascual A, Preat T. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- 41.McGuire SE, Le PT, Davis RL. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 42.Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 44.Rubin GM, Spradling AC. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 45.Guo HF, Tong J, Hannan F, Luo L, Zhong Y. Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- 46.O'Connell PO, Rosbash M. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.