Abstract
mRNA of κ opioid receptor (KOR) can be transported to nerve fibers, including axons of dorsal root ganglia (DRG), and can be locally translated. Yeast three-hybrid screening identifies Copb1 as a kor mRNA-associated protein that form complexes with endogenous kor mRNA, which are colocalized in the soma and axons of DRG neurons. Axonal transport of kor mRNA is demonstrated, directly, by observing mobilization of biotin-labeled kor mRNA in Campenot chambers. Efficient transport of kor mRNA into the side chamber requires Copb1 and can be blocked by a drug that disrupts microtubules. The requirement for Copb1 in mobilizing kor mRNA is confirmed by using the MS2–GFP mRNA-tagging system. Furthermore, Copb1 also facilitates the translation of kor mRNA in the soma and axons. This study provides evidence for a microtubule-dependent, active axonal kor mRNA-transport process that involves Copb1 and can stimulate localized translation and suggests coupling of transport and translation of mRNAs destined to the remote areas such as axons.
Keywords: dorsal root ganglia neurons
Three major opioid receptor types, μ, δ, and κ, are known (1, 2), each interacting with specific ligands to modulate pain sensation, consciousness, and autonomic functions. Pharmacological studies have demonstrated the significance of the number (3) of opioid receptors and their subcellular distribution (4–7) in the manifestation of opioid drugs. Studies of gene regulation have revealed that although transcriptional regulation is critical for cells to express opioid receptor mRNAs (8), these mRNAs are mostly silent and require extensive posttranscriptional regulation for the ultimate production of these proteins (9). Using the κ opioid receptor (KOR) as a model, we have begun to uncover several posttranscriptional regulatory events important for the expression of KOR protein. These include alternative mRNA splicing, changing mRNA stability (10), localized translation (11), and mRNA transport (12, 13) in neurons.
Active transport of RNAs in extensively compartmentalized cells has been established, but for mammalian neurons, it has remained a highly debated issue whether this type of transport process occurs, or is required, for axonal proteins detected in the sensory neurons (14). “Neuronal RNA granules” have been implicated to play roles in various important processes including sorting/storing mRNAs, degrading mRNAs, translational control, and mobilizing mRNAs (15–18). Most of these studies focus on components transported to the dendrites, such as Staufen-containing RNA granules (19, 20) and CaMKIIα mRNA (21, 22). Recently, a thoroughly examined transport process, mobilization of actin mRNA involving a specific zip-code-binding protein ZBP1, has been found also to occur in developing axons (23, 24). In terms of the motor, or machinery, for these transport processes, studies have suggested that microtubule is important and kinesin and dynein are involved (25). With the increasingly appreciated role for RNA transport in neurons, an active RNA transport process for delivering certain mRNAs, including mRNAs for nonstructural components, to the axonal compartments of sensory neurons has been speculated.
It is known that KOR protein can be detected in sensory neurons both pre- and postsynaptically (4, 5). Although presynaptic KOR can be derived from proteins that are synthesized in soma and transported by vesicles (26, 27), they also may be produced locally from mRNAs located in these areas where endogenous kor mRNAs can be detected and translated (12). Recently, we were able to demonstrate axonal transport and translation of kor mRNA in neurons including dorsal root ganglia (DRG) neurons, a unique example of axonal transport of mRNA encoding a nonstructural component (12). However, it was not yet clear whether this indeed was an active transport process and what machinery or components were required for this process. To shed light on the nature of, and the transport machinery for, mobilizing kor mRNAs to axons, we launched a yeast three-hybrid screening experiment to identify proteins associated with the UTRs of kor mRNA. From these screening experiments, multiple clones of Copb1 (coatomer protein complex, subunit beta 1), a subunit found in the COP1 vesicle coatomer complexes and the clathrin-binding complexes (28, 29), were repeatedly obtained by using either the 5′- or the 3′-UTR of kor mRNA as the probe. We now present data to demonstrate that Copb1 is associated with the endogenous kor mRNA and its complex contains, at least, the RNA-binding protein Hu antigen R (HuR) and a kinesin motor subunit. By using the MS2–GFP tagging system and Campenot chamber device, we also present evidence that Copb1 plays a functional role in facilitating the transport of kor mRNAs into axons of DRG neurons. We further demonstrate that this is a microtubule-dependent active transport process, and that Copb1 also plays a novel role in facilitating translation of kor mRNA in cortical neurons and enhances translation of proteins in the axonal compartment of DRG neurons.
Results
Physical Association of Copb1 with kor mRNA, Kinesin, and HuR.
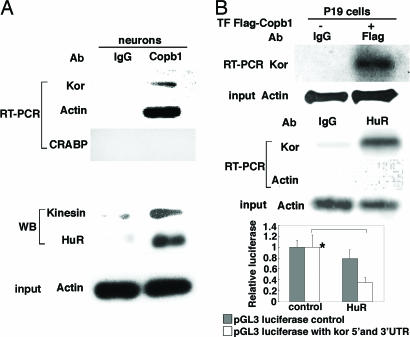
We have reported previously that kor mRNA can be detected and transported into the axons of DRG neurons and that, in these neurons, ≈16% of total kor mRNA is localized in the axons (12). To identify kor mRNA-binding proteins that might be involved in its transport, we then conducted yeast three-hybrid screening experiments by using either the 5′- or the 3′-UTR of kor mRNA (10) as the probe to screen mouse brain cDNA libraries. Independently, multiple clones of Copb1 were obtained from both experiments. We then examined the physical association of Copb1 with endogenous kor mRNA in primary cortical neurons by using coimmunoprecipitation, which clearly detected endogenous kor mRNA coprecipitated with anti-Copb1, as shown in the upper three gels of Fig. 1. Physical association of Copb1 with endogenous kor mRNA was validated in a Flag–Copb1-transfected cell line that is known to expresses abundant kor mRNA, P19 (Fig. 1B, upper gels). The possibility of direct interaction of Copb1 with kor mRNA was examined by using a GST-tagged Copb1 to determine whether Copb1 could directly pull down in vitro-synthesized kor mRNA. This in vitro direct protein–RNA interaction test failed to detect direct interaction between Copb1 and kor mRNA (data not shown), suggesting that kor mRNA is indirectly associated with Copb1-containing complexes through other components. The Copb1 complex was then examined to identify other associated proteins that might be involved in mobilizing or stabilizing kor mRNA. It appeared that the Copb1 complex also contained the kinesin motor and an RNA-binding protein, HuR, that is known to stabilize mRNA and repress translation (30, 31) (Fig. 1A, “WB” and “input” gels). Association of HuR with kor mRNA was further supported, as demonstrated in an RNA–protein coprecipitation experiment (Fig. 1B, middle gels). Furthermore, HuR exerted a repressive activity on translation of kor mRNA, as revealed in the translational reporter assay (graph in Fig. 1B). These results indicate that Copb1, HuR, and kinesin form a stabilizing complex, or granule, that mobilizes kor mRNA through, at a minimum, the kinesin motor. As expected, HuR indeed repressed KOR translation.
Fig. 1.
In vivo association of Copb1 with kor mRNA, HuR, and kinesin. (A) RNA immunoprecipitation (Upper) and protein coimmunoprecipitation (Lower) of anti-Copb1 precipitated complexes from cortical neurons. In A Upper, RT-PCR was used to detect kor transcript in Copb1 complex with cellular retinoic-acid-binding protein (CRABP) mRNA as a negative control. In A Lower, Western blot (WB) was used to detect proteins associated with Copb1 and the input (actin). (B) kor mRNA immunoprecipitated with Copb1 (Flag-Copb1) (Top) and HuR (Middle) from P19 cells. Input (actin) is also shown. For the graph (Bottom), a reporter assay was used to show the effect of HuR on translation of kor mRNA in P19. *, P <0.05 comparing with and without HuR overexpression in pGL3 luciferase with 5′- and 3′-UTR.
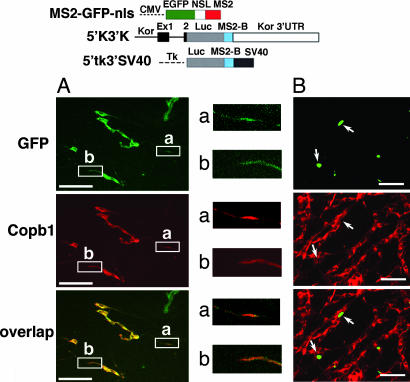
To validate in vivo association of kor mRNA with Copb1, we used an MS2–GFP tagging system to examine whether the MS2–GFP-tagged kor mRNA could be colocalized with endogenous Copb1 (detected by anti-Copb1). As shown in Fig. 2A Upper and Middle, the WT intact kor mRNA tagged with MS2–GFP (5′k3′k) was clearly transported to the extended axons of these DRG neurons and could be colocalized with endogenous Copb1 (Fig. 2A Middle and Lower). To the contrary, a negative control mRNA with the same MS2–GFP sequence tagged to SV40 RNA sequences (5′tk3′SV40) remained exclusively in the soma (Fig. 2B). This result further supports that kor mRNA indeed can form a complex with Copb1, and this can occur in the extended axons of DRG neurons.
Fig. 2.
Colocalization of Copb1 with kor mRNA in DRG neurons. The constructs of the GFP–MS2-tagging system are shown at the top. NSL, nuclear localization signal. (A) The left sides, of the gels show the culture harboring the intact kor mRNA tagged with GFP–MS2, 5′k3′k. GFP (green) signifies the location of kor mRNA, whereas Copb1 (red) depicts the distribution of endogenous Copb1. Overlapped images are shown at the bottom. The right sides of the gels are enlarged images of two boxed areas containing axons. (B) Images of the culture harboring the negative control 5′tk3′SV40. a and b are enlarged ×3. (Scale bars, 100 μm.)
A Functional Role for Copb1 in Mobilizing kor mRNA to Axons of DRG Neurons.
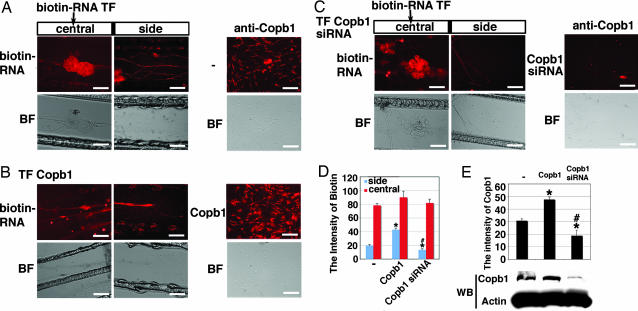
Copb1 could be found in the COP1 vesicle coat, known for transporting proteins or in the clathrin-binding complexes (28, 29). The result that kor mRNA could be associated with Copb1 suggested an interesting possibility that kor mRNA transport might have involved a machinery, or granule, that contained Copb1. To test this possibility, we exploited an RNA-transfection procedure to directly monitor the movement of labeled mRNA in combination with the use of a compartment culture device, a Campenot chamber, where soma and axons can be separated in living cultures to be individually manipulated and monitored. Gain- and loss-of-function studies were then conducted to determine the functionality of Copb1 in DRG neurons cultured in the central compartment of Campenot chambers by monitoring mobilization (into the side compartment) of biotin-labeled kor mRNA that was introduced only into the central compartment. As shown in Fig. 3, for control DRG neurons, biotin-labeled kor mRNA was effectively transported to the side chamber as predicted (Fig. 3A), which was enhanced when the DRG neurons received supplemented Copb1 by transfection (Fig. 3B). Importantly, for the DRG culture that received siRNA of Copb1, transport of biotin-labeled kor mRNA to the side chamber was significantly reduced (Fig. 3C). The efficiency of RNA transport was quantified by comparing the intensity of biotin label in the central and side chambers for each group. As shown in Fig. 3D, both the enhancement (due to elevating Copb1 level) and the suppression (due to Copb1 knockdown) of axonal kor mRNA transport in DRG neurons are statistically significant. The predicted expression pattern of Copb1 was confirmed in immunohistochemistry and Western blot shown in Fig. 3E.
Fig. 3.
The role of Copb1 in kor mRNA transport in DRG neurons. (A) Biotin-labeled kor mRNA was introduced into the central compartment of the Campenot slide, and its distribution in the central and the side chambers was monitored (two leftmost panels). The expression of endogenous Copb1 was monitored by anti-Copb1 immunohistochemistry (rightmost panel). (B) A similar experiment as that described for A that was conducted in the culture transfected with Copb1 for 24 h. The elevation of Copb1 was confirmed by immunohistochemistry in the rightmost panel. (C) A similar experiment as that described for A was conducted in the culture receiving Copb1 siRNA for 24 h. Knockdown of Copb1 was also monitored by immunohistochemistry (rightmost panel). Both biotin and anti-Copb1 signals are shown in red. BF, bright field; TF, transfection. (D) A statistical analysis of the amounts of biotin-labeled RNA in the central and the side chambers (n = 3; *, P < 0.025 for comparing to the control; #, P < 0.025 for comparing the Copb1–siRNA culture to the Copb1-transfected culture). (E) The graph shows the statistical analysis of the intensity of Copb1 for each group (n = 5; *, P < 0.025 for comparing with the control; #, P < 0.025 for comparing the Copb1–siRNA culture with the Copb1-transfected culture). The blot shows Western blot results detecting Copb1 and actin expression. (Scale bars, 50 μm.)
Microtubule-Dependent Axonal Transport of kor mRNA in DRG Neurons.
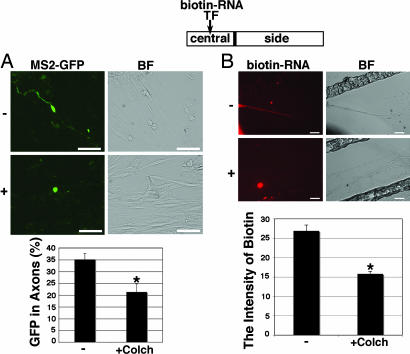
The finding that Copb1 was critical for the mobilization of kor mRNA to the axons of DRG neurons suggested that kor mRNA transport might depend on microtubules because the transport of Copb1-containing vehicles was known to depend on microtubules (32, 33). We then applied a drug, colchicines, that could disrupt microtubules to test whether kor mRNA transport could be affected by this drug (Fig. 4). As shown in Fig. 4A, by using the MS2–GFP tagging system, we found that mobilization of MS2–GFP-tagged kor mRNA was dramatically ablated when the culture was exposed to this drug. By observing biotin-labeled kor mRNA introduced into the central compartments of Campenot chambers (Fig. 4B), we also detected a dramatic reduction in the detectable biotin-labeled kor mRNA in the side compartment when colchicine was added to the culture, supporting that transport of kor mRNA to axons was indeed suppressed by the drug that disrupted microtubules. The effect of colchicine on neuron survival in the experimental condition was evaluated based on the morphology of DRG neurons [supporting information (SI) Text]; the expression of a growth associated protein (GAP43) and cell survival counts before and after colchicine treatment (SI Fig. 6). The result showed that colchicine did not significantly affect neuronal viability under the experimental condition (10 μg/ml for 1 h), although cells seemed to be morphologically unhealthy after a prolonged treatment, such as a 2-h treatment (data not shown). Together, these results indicate that axonal kor mRNA transport involves an active mechanism (such as via the kinesin motor) that depends on Copb1-containing complexes or granules moving along the microtubules.
Fig. 4.
kor mRNA transport is microtubule dependent. (A) Micrographs show the mobilization of MS2–GFP-tagged kor mRNA (Left) and the negative control (Right) in DRG neurons with (Lower) or without (Upper) the microtubule disruption drug colchicines. (B) Micrographs show mobilization of biotin-labeled kor mRNA (Left) from the central to the side chambers of the Campenot cultures with (Lower) or without (Upper) colchicines after Copb1 transfection for 24 h to stimulate the transport. Only the side chambers are shown. GFP signals are in green, and biotin-RNA signals are in red. (Scale bars, 25 μm.) Statistical analyses is shown in the graphs (for both, n = 3; *, P < 0.025 for comparing with the control). BF, bright field.
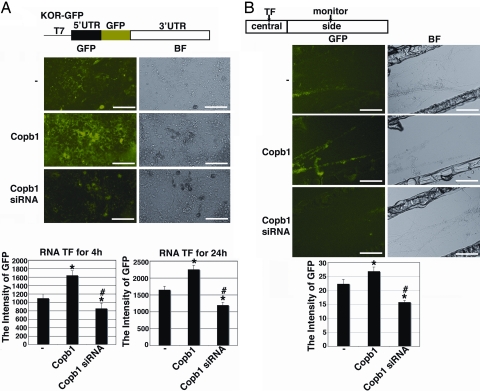
Copb1-Facilitated Axonal kor mRNA Translation in DRG Neurons.
Previously, we have demonstrated that kor mRNA directly introduced into the axonal compartment of DRG neurons could be locally translated (12). Given that kor mRNA mobilization from the soma to the axon could be enhanced by elevating the level of Copb1, and knockdown of Copb1 drastically hinders this transport process, it was important to determine whether this RNA transport process might also be involved in, or coupled to, local translation of the target mRNA. To examine this possibility, we used a fusion transcript (KOR–GFP) that contained the intact 5′- and 3′-UTRs of kor mRNA with its entire coding region replaced with a GFP coding sequence. Thus the translation of this fusion transcript (monitored by GFP expression) that is under the control of kor mRNA signals, could be followed in living cells in real time. The effect of Copb1 on local KOR translation, an effect that may be coupled to the mRNA transport of KOR, could then be monitored, as shown in Fig. 5. Clearly, in the cortical neuron culture receiving supplemented Copb1, GFP expression was significantly elevated as early as 4 h after transfection (Copb1-labeled row in Fig. 5A). Interestingly, GFP expression in neurons with Copb1 knockdown was significantly lowered (Copb1 siRNA-labeled row in Fig. 5A), suggesting that Copb1 could enhance the translation of KOR transcripts. To further examine whether this might be due to coupling of kor mRNA translation with its transport, the experiment was repeated by using DRG cultures on the Campenot chambers (Fig. 5B). Interestingly, in the culture supplemented with Copb1 and receiving KOR–GFP transcript only in the central chamber, the expression of GFP in the side chamber was significantly elevated within 24 h (Copb1-labeled row in Fig. 5B), whereas in the culture with Copb1 knockdown, the expression of GFP in the side chamber was almost invisible at this time (Copb1 siRNA-labeled row in Fig. 5B). These results revealed an interesting “coupling” phenomenon of mRNA translation and its transport. The effect of Copb1 on KOR translation (GFP expression), as detected by using the conventional culture (Fig. 5A) or the Campenot chamber device (Fig. 5B), was found to be significant, as shown in the graphs.
Fig. 5.
Copb1-stimulated kor mRNA translation. (A) The construct of kor template (KOR–GFP) with its coding sequence replaced, in-frame, with the GFP coding sequence is shown above the micrographs. The micrographs show in vitro synthesized, capped, and polyadenylated KOR–GFP transcript introduced into primary cortical neurons of a control culture (Top), a Copb1-transfected culture (Middle), and a culture receiving Copb1 siRNA (Bottom). GFP signals were monitored at 4 and 24 h, and the statistical analyses are shown in the graphs. (B) The diagram above the photographic images represents the central and side compartments of a Campenot chamber. Micrographs show KOR–GFP transcript introduced into the central compartment of the Campenot culture of a control (Top), a culture receiving supplemented Copb1 (Middle), or a culture receiving Copb1 siRNA (Bottom). Only the side chambers are shown. The GFP signals in the side chamber were monitored at 24 h, as shown, and the statistical analysis is shown in the graph. GFP signals are in green (n = 5, *, P < 0.025 for comparing with the control; #, P < 0.025 for comparing Copb1 siRNA with Copb1 transfection). (Scale bars, 50 μm.)
Discussion
We demonstrated previously that axonal transport of mRNA encoding nonstructural components, such as KOR, could occur in sensory neurons, including DRG neurons (12). However, it was unclear if this observation was mediated by an active transport process and what machinery was involved or required. The current study illustrates that this indeed is an active transport process that depends on microtubules and involves at least the kinesin motor, and the study identifies an important component required for this RNA transport process that, surprisingly, is a subunit found in the COP1 vesicles, Copb1. Our data further suggest a “coupling” mechanism that potentially can link active, or possibly stimulated, mRNA transport process with translational activation.
By using direct protein–RNA interaction tests, we failed to detect direct interaction of kor mRNA with Copb1. This would indicate that kor mRNA is carried by, or tethered to, the transport complex, or granule, through other RNA-binding proteins. Interestingly, an RNA-binding protein that is known to stabilize and repress mRNA, HuR, was also found in the Copb1 complex, and HuR was confirmed to be associated with kor mRNA and repress translation of kor mRNA (Fig. 1). This suggests that Copb1 might become associated with kor mRNA through HuR, which can possibly bind multiple target sites in the 3′-UTR of kor mRNA (data not shown). We attempted to determine whether any fraction of the endogenous kor mRNA pool could be enclosed in any form of vesicles, including COP1 vehicles, by examining whether kor mRNA that associated with vesicles prepared from mouse brain tissues could be protected from harsh RNase treatment. It appeared that after RNase treatment, kor mRNA in the vesicle preparation was completely degraded (data not shown). This indicates that kor mRNA is most likely tethered to the RNA granules containing Copb1 but not enclosed in the vesicles. Because Copb1 was cloned from the three-hybrid screening by using either the 5′- or the 3′-UTR of kor mRNA, and its complex contained HuR for which multiple binding sites were predicted within the 3′-UTR of kor mRNA, Copb1 was presumably associated with the 3′-UTR of kor mRNA through HuR. The proteins mediating the association of Copb1 with the 5′-UTR of kor mRNA remain to be identified. To this end, it is surprising that the 5′-UTR kor mRNA-binding protein Grb7 (34) was not detected in the Copb1 complex (see below).
Previously, we have found that most kor mRNA is repressed until it is stimulated for translation (11, 34). It has also been suggested that mRNAs, during transport, are usually silenced by repressors (35–37). The recently identified translational repressor of kor, Grb7, can bind to the 5′-UTR of kor mRNA via a stem–loop (34), but it seems to be missing from the Copb1 complex in the cortical neuron preparation used in experiments of Fig. 1 (not shown). The silencing of kor mRNA during transport is probably mediated by a different repressive mechanism. For instance, HuR may perform such a function. Multiple HuR-interacting domains are predicted, scattering around the 3-kb-long 3′-UTR of kor mRNA. Quite likely, kor mRNA can be associated with, and stabilized by, HuR via multiple contact points. However, we have not ruled out other unknown factors possibly involved in translational repression of kor mRNA during transport.
Interestingly, elevating the level of Copb1 to enhance the transport of kor mRNA also facilitates its translation, and knockdown of Copb1, which is predicted to hinder the transport of kor mRNA, suppresses its translation. This holds true not only for the soma but also for axons when the transcripts are originated from the soma (Fig. 5). In a parallel experiment shown in Fig. 5B, in which the KOR–GFP transcript was introduced directly into the side chamber of the culture that received either supplemented Copb1 or its siRNA, the expression of GFP in the side chamber seemed to be unaffected (data not shown). This suggests that translation of kor mRNA in axons is enhanced because more soma-originated transcripts are mobilized to these areas. Thus, “coupling” of the mRNA-transport process with axonal translation seems to be mediated by events initiated from the soma, at least for kor mRNA in the DRG neurons. Once mRNA reaches its destination (such as already in the axon), its translation is no longer affected by Copb1-mediated transport. Presumably for efficient translational activation, kor mRNA must be located, or delivered, to certain areas via Copb1 complexes or granules starting from the soma. These granules then interact with certain translational regulators (or activators) once they reach final target sites. For those transcripts already at the target sites, their translation is no longer stimulated by the transport process. It would be important to identify the type of translational regulators that can interact with this type of RNA-transport complexes or granules. For instance, it would be crucial to determine whether translational repression is switched off and replaced with an activating component once the RNA reaches certain target sites. Alternatively, the transported mRNA simply may be released from the granules that are translationally repressive to certain active domains of translation. In either scenario, it is predicted that this type of Copb1-containing RNA granules or complexes must dynamically interact with various translational regulators.
Copb1 may be one of the seven subunits of COP1 coatomer complex or clathrin-binding complex that are known for transporting proteins among plasma membrane, trans-Golgi network, endoplasmic reticulum, and the lysosome/vacuole (28, 29). Mobilization of these vesicles or complexes is known to depend on motors such as kinesin and dynein families (27, 38) that move along microtubules. The drug that disrupts microtubules also blocks kor mRNA transport, supporting that kor mRNA transport is mediated by a transport complex similar to either the COP1 vesicle or the clathrin-binding complex. This is consistent with the data showing the association of kinesin with the Copb1/HuR complex (Fig. 1). However, the interaction of the kinesin motor with the kor mRNA/Copb1 complex awaits further investigation. The possibility of the dynein motor being involved in this process also requires further studies in the future.
Materials and Methods
The experiments involving animals were approved by the Institutional Animal Care and Use Committee of the University of Minnesota. All of the experiments conformed to regulatory standards.
Plasmid Constructs, Yeast Three-Hybrid Screening, and siRNAs.
The plasmids for the single-stranded RNA phage capsid protein MS2-fused nuclear GFP (MS2–GFP–nuclear localization signal), the MS2-binding-site-tagged kor mRNA (5′K3′K), the MS2-site-tagged negative control mRNA (5′tk3′SV40), and the KOR–GFP fusion were as described previously (12, 13). Flag-tagged Copb1 was made by inserting the full-length Copb1 cDNA amplified with a primer pair (5′-GCG AGA TCT ACG GCA GCC GAG AAC GTG TGC-3′ and 5′-GCT CTA GAT TAT AGA CTA GTC TTC TTT TG-3′) into BglII/XbaI sites of a Flag-tag expression vector pCMX. The screening of kor mRNA-binding proteins was performed by using a RNA–protein hybrid Hunter kit (Invitrogen, Carlsbad, CA) as described previously (34). Two small Copb1 siRNAs were purchased from Qiagen (Valencia, CA). The target sequence was TAC GTT AAT TAA CGT GCC AAT (184–204 region) and GAC CAA GAT TGC ATT ACG CTA (1840–1860 region). The transfection of Copb1 siRNA was conducted by using a HiPerFect transfection reagent according to the procedure recommended by the manufacturer (Qiagen, Santa Clarita, CA). Briefly, 37.5 ng of siRNA was diluted in 100 μl of DRG culture medium without serum. A total of 3 μl of HiPerFect transfection reagent was added to the diluted siRNA. To form complexes, the mixture was incubated for 5–10 min at room temperature. The complexes were added to cells for 24–48 h before biotin–RNA transfection and were continuously incubated for another 24 h before fixing the cells.
RNA Immunoprecipitation and RT-PCR.
Immunoprecipitation was modified from a previously described protocol (39). Mouse primary cortical neurons or transfected P19 cells were treated with 1% formaldehyde at room temperature for 10 min, and cross-linking was stopped by glycine (pH 7.0, 0.25 M). The samples were washed twice and resuspended in 1.5 ml of radioimmunoprecipitation assay buffer [50 mM Tris·HCl (pH 7.4)/1% Nonidet P-40/0.5% sodium deoxycholate/0.05% SDS/1 mM EDTA/150 mM NaCl] containing protease inhibitors and 50 units/ml RNase inhibitor. The suspension was sonicated, centrifuged to remove insoluble materials, and precleared by incubating with 20 μl protein G-agarose bead and 100 μl/ml Escherichia coli tRNA. Aliquots of the precleared supernatants were incubated with 1 μl of anti-Copb1 anti-kinesin, anti-actin, or IgG, and 25 μl of protein G beads at 4°C for 4 h. The beads were collected and washed with radioimmunoprecipitation assay buffer at alternating high-salt (1 M NaCl) and low-salt (150 mM NaCl) conditions and suspended in 200 μl of 50 mM Tris·Cl (pH 7.0)/5 mM EDTA/10 mM DTT and incubated at 70°C for 45 min to reverse cross-linking. The immunoprecipitated RNA was extracted with an equal volume of phenol (pH 4.3) followed by RT-PCR with the OneStep RT-PCR kit (Qiagen). The kor cDNA was amplified by using the primer pair 5′-CATCATCAGGAAACTGCA-3′ and 5′-TGGTCATGTTTGTCATC-3′, followed by Southern blots as described previously (10).
Reporter Assay.
Reporter assay was conducted as described (11). In brief, P19 cells were transfected with a pGL3 vector or a KOR translational reporter (11) together with the HuR expression plasmid and an internal control, LacZ. Thirty hours later, transfected cells were collected, and specific reporter activities were determined by normalizing to the internal control LacZ. Statistical analysis was performed by using Student's t test.
Neuron Cultures, Western Blot, and Immunohistochemistry.
Procedures for mouse cortical neuron culture (40), rat DRG neuron culture (12, 13), and Western blot (11) were as described previously. Transfection was conducted by using a Lipofectamine 2000 kit (Invitrogen). Immunohistochemistry was conducted as described (12). Images were taken by using an inverted fluorescence microscope (Nikon, Tokyo, Japan) and quantified with the help of ImageJ software (National Institutes of Health, Bethesda, MD) as described previously (12). For quantification, three to five individual axons or random areas were measured for each treatment, and the experiment result was analyzed by the paired Student's t test. Colchicines treatment was conducted by using 10 μg/ml colchicines for 1 h (Sigma, St. Louis, MO).
Compartment Culture, in vitro Transcription, and RNA Transfection.
Rat DRG neuron compartment culture preparation, in vitro transcription, and RNA transfection were as described previously (12, 41, 42).
Supplementary Material
Acknowledgments
We thank our colleagues Drs. X. Hu and G. Li for helping with the three-hybrid screening. This work was supported by National Institutes of Health Grants DA11190 (to L.-N.W.), DK54733 (to L.-N.W.), DK60521 (to L.-N.W.), K02 DA13926 (to L.-N.W.), DA00564 (to H.H.L.), DA01583 (to H.H.L.), DA11806 (to H.H.L.), and K05-DA70554 (to H.H.L.).
Abbreviations
- DRG
dorsal root ganglia
- HuR
Hu antigen R
- KOR
κ opioid receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703805104/DC1.
References
- 1.Goldstein A, Naidu A. Mol Pharmacol. 1989;36:256–272. [PubMed] [Google Scholar]
- 2.Pasternak GW. Neuropharmacology. 2004;47(Suppl 1):312–323. doi: 10.1016/j.neuropharm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Law PY, Erickson LJ, El-Kouhen R, Dicker L, Solberg J, Wang W, Miller E, Burd AL, Loh HH. Mol Pharmacol. 2000;58:388–398. doi: 10.1124/mol.58.2.388. [DOI] [PubMed] [Google Scholar]
- 4.Shuster SJ, Riedl M, Li XR, Vulchanova L, Elde R. J Neurosci. 1999;19:2658–2664. doi: 10.1523/JNEUROSCI.19-07-02658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoffelmeer AN, Rice KC, Jacobson AE, Van Geleren JG, Hogenboom F, Heijna MH, Mulder AH. Eur J Pharmacol. 1988;154:169–178. doi: 10.1016/0014-2999(88)90094-5. [DOI] [PubMed] [Google Scholar]
- 6.Sauliere A, Gaibelet G, Millot C, Mazeres S, Lopez A, Salome L. FEBS Lett. 2006;580:5227–5231. doi: 10.1016/j.febslet.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 7.Marie N, Aguila B, Allouche S. Cell Signal. 2006;18:1815–1818. doi: 10.1016/j.cellsig.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Wei L-N, Loh HH. Curr Opin Pharmacol. 2002;2:69–75. doi: 10.1016/s1471-4892(01)00123-0. [DOI] [PubMed] [Google Scholar]
- 9.Wei L-N, Law YP, Loh HH. Front Biosci. 2004;9:1665–1679. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei L-N, Hu X, Bi J, Loh HH. Mol Pharmacol. 2000;57:401–408. [PubMed] [Google Scholar]
- 11.Tsai NP, Bi J, Loh HH, Wei L-N. J Neurosci. 2006;26:9743–9749. doi: 10.1523/JNEUROSCI.3014-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi J, Tsai NP, Lin YP, Loh HH, Wei L-N. Proc Natl Acad Sci USA. 2006;103:19919–19924. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi J, Hu X, Loh HH, Wei L-N. Mol Pharmacol. 2003;64:594–599. doi: 10.1124/mol.64.3.594. [DOI] [PubMed] [Google Scholar]
- 14.Job C, Eberwine J. Nat Rev Neurosci. 2001;2:889–898. doi: 10.1038/35104069. [DOI] [PubMed] [Google Scholar]
- 15.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 16.Tongiorgi E, Righi M, Cattaneo A. J Neuronsci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krichevsky AM, Kosil SK. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 18.Tiruchinapalli MD, Oleynikov Y, Kelic S, Shenoy MS, Hartley A, Stanton KP, Singer RH, Bassell GJ. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 20.Mallardo M, Deitiqhoff A, Muller J, Goetze B, Macchi P, Peter C, Kiebler MA. Proc Natl Acad Sci USA. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rook MS, Lu M, Kosil KS. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori Y, Imaizumi, Katayama T, Yoneda T, Tohyama M. Nat Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- 23.Ross AF, Oleynikov Y, Kislauskis EH, Tanejs KL, Singer RH. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 25.Hirokawa N. J Neurosci. 2006;26:7139–7142. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malkus P, Jiang F, Schekman R. J Cell Biol. 2002;159:915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 28.Sytnyk V, Leshchyns'ka I, Dityatev A, Schachner M. J Cell Sci. 2004;117:381–388. doi: 10.1242/jcs.00956. [DOI] [PubMed] [Google Scholar]
- 29.Nickel W, Brugger B, Wieland FT. J Cell Sci. 2002;115:3235–3240. doi: 10.1242/jcs.115.16.3235. [DOI] [PubMed] [Google Scholar]
- 30.Myer VE, Fan XC, Steitz JA. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorospe M. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- 32.Nakata T, Hirokawa N. J Cell Biol. 2003;162:1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton AC, Ehlers MD. Nat Cell Biol. 2004;6:585–591. doi: 10.1038/ncb0704-585. [DOI] [PubMed] [Google Scholar]
- 34.Tsai NP, Bi J, Wei L-N. EMBO J. 2007;26:1522–1531. doi: 10.1038/sj.emboj.7601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH. Mol Cell Biol. 2006;26:277–292. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 37.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 38.Chen JL, Fucini RV, Lacomis L, Erdjument-Bromage H, Tempst P, Stamnes M. J Cell Biol. 2005;169:383–389. doi: 10.1083/jcb.200501157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB. Proc Natl Acad Sci USA. 2003;100:15566–15571. doi: 10.1073/pnas.2536695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang FJ, Nutter LM, Thayer SA. Biochem Pharmacol. 1997;54:181–187. doi: 10.1016/s0006-2952(97)00158-5. [DOI] [PubMed] [Google Scholar]
- 41.Kimpinski K, Campenot RB, Mearow K. J Neurobiol. 1997;33:395–410. doi: 10.1002/(sici)1097-4695(199710)33:4<395::aid-neu5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 42.Ure DR, Campenot RB. J Neurosci. 1997;17:1282–1290. doi: 10.1523/JNEUROSCI.17-04-01282.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.