Abstract
Hibernation is a fascinating, yet enigmatic, physiological phenomenon during which body temperature and metabolism are reduced to save energy. During the harsh season, this strategy allows substantial energy saving by reducing body temperature and metabolism. Accordingly, biological processes are considerably slowed down and reduced to a minimum. However, the persistence of a temperature-compensated, functional biological clock in hibernating mammals has long been debated. Here, we show that the master circadian clock no longer displays 24-h molecular oscillations in hibernating European hamsters. The clock genes Per1, Per2, and Bmal1 and the clock-controlled gene arginine vasopressin were constantly expressed in the suprachiasmatic nucleus during deep torpor, as assessed by radioactive in situ hybridization. Finally, the melatonin rhythm-generating enzyme, arylalkylamine N-acetyltransferase, whose rhythmic expression in the pineal gland is controlled by the master circadian clock, no longer exhibits day/night changes of expression but constantly elevated mRNA levels over 24 h. Overall, these data provide strong evidence that in the European hamster the molecular circadian clock is arrested during hibernation and stops delivering rhythmic output signals.
Keywords: deep torpor, suprachiasmatic, euthermia
In mammals, the hibernation season consists of recurring torpor bouts of low body temperature (Tb) alternating with short periods of euthermia during which Tb returns to 37°C (1–5). During deep torpor, energetically expensive cellular processes such as transcription and translation are severely depressed, and physiological functions such as heart rate, respiration, immune and renal functions, and neural activity run at greatly reduced rates (1, 2). The circadian system temporally coordinates internal biological processes with each other and with the environment to ensure health and survival (6–8). Circadian rhythms are temperature-compensated and run at a relatively constant pace under various temperatures (6). However, it has long been debated whether the circadian system remains functional during deep hibernation (3, 5, 9). Although several studies have addressed this question (10–22), uncertainty prevails because clock outputs rather than the core clockwork machinery were examined. In mammals, the master circadian clock is located in the suprachiasmatic nucleus of the hypothalamus (SCN) (6–8). Circadian oscillations within the SCN neurons result from the recurrent expression of so-called clock genes that interact in complex, interlocked transcription/translation feedback loops (6, 7). Assessing such molecular oscillations in the SCN of hibernating animals may be a valuable approach to readdressing the issue of the circadian clockwork function under deep torpor.
Results
European Hamsters Display a Characteristic Hibernation Pattern.
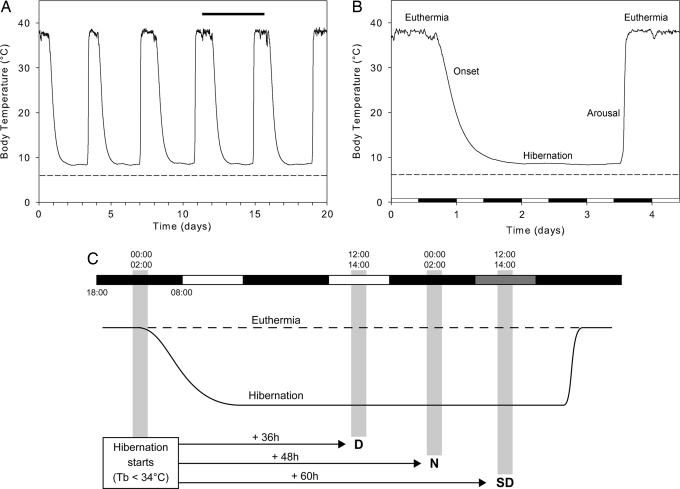
For our investigations, we used the European hamster (Cricetus cricetus L.), a well defined hibernator (14, 21, 23, 24). Hamsters raised outdoors were transferred in September to a climatic room kept at 6 ± 2°C under short photoperiodic regimen, and individual Tbs were recorded by a telemetric system. In these conditions, comparable with natural conditions in December, the bouts of deep torpor characteristic of the hibernation cycle were expressed regularly (Fig. 1A) (21, 24). During deep torpor, the metabolic rate is typically reduced to 2–4% of euthermic rates, and Tb actively drops to approach ambient temperature (Ta) (1, 2). In this experiment, the Tb of hibernating hamsters was 8–10°C (Fig. 1B).
Fig. 1.
European hamster's hibernation pattern and experimental paradigm. (A) Typical Tb recording (animal B17) illustrating the major phases of the hibernation cycle of the European hamster. Three weeks of recording (November 25 to December 14, 2005) show five hibernation cycles. The ambient temperature (Ta) of the climatic room was set at 6 ± 2°C (dashed line). The horizontal black bar represents the period detailed in B. (B) Focus on a single hibernation cycle (December 6 to December 10, 2005). Entrance into hibernation (Tb < 34°C) occurred during nighttime. (C) Experimental paradigm used for the experiment. For the euthermic group, the hamsters had a Tb close to 37°C and were aroused from hibernation for >48 h. These animals were killed at mid-day (D, 1300; n = 6), mid-night (N, 0100; n = 6), or subjective mid-day (SD, 1300; darkness from the previous day; n = 5). For the hibernating group, only the animals entering torpor (Tb <34°C) between 0000 and 0200 (mid-night) were considered. Their killing occurred 36 h (day; n = 4), 48 h (night; n = 5), or 60 h (subjective day; no lights on for the last day; n = 7) later. Light and dark bars indicate day and night, respectively; dark gray bar, subjective day.
The European hamster enters hibernation preferentially in the middle of the night (14, 21). We took advantage of this characteristic to design our experiment, as illustrated in Fig. 1C. Radioactive in situ hybridization was used to assess gene expression in the SCN of euthermic versus hibernating hamsters, killed during daytime (D), nighttime (N), or during the subjective day (SD). Having all hibernating animals entering hibernation at the same circadian phase (middle of the night) allowed comparison of circadian gene expression among them as well as with euthermic hamsters.
Effect of Deep Torpor on Circadian Clock Gene Expression.
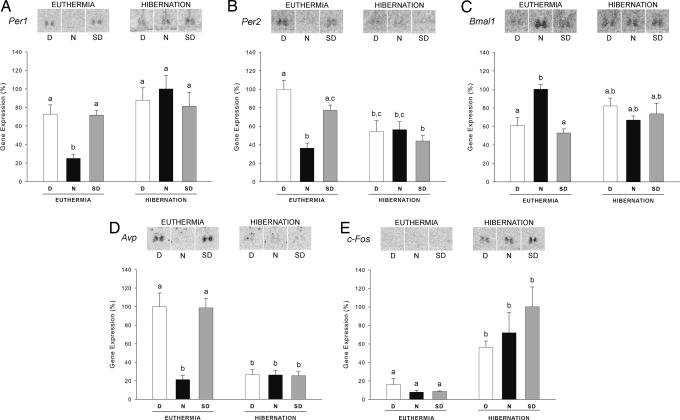
In the molecular core of the clock, the transcription factors CLOCK and BMAL1 drive the expression of the Period (Per) and Cryptochrome (Cry) genes, and in turn PER and CRY proteins heterodimerize and repress CLOCK/BMAL1-driven transcription (6, 7). To compare the state of these molecular loops during deep torpor versus euthermia, we examined the expression of three major clock genes: Per1, Per2, and Bmal1 (Fig. 2 A–C, respectively). As expected in euthermic animals, significant day/night changes of expression were observed, with high expression of Per1 and Per2 during the day and the subjective day (Per1 and Per2, P < 0.001; N versus D and SD), and high expression of Bmal1 during nighttime (Bmal1, P = 0.008; N versus D and SD). In contrast to euthermic animals, no day/night change of expression occurred in the SCN of hibernating hamsters (Fig. 2 A–C, respectively; Per1, Per2, and Bmal1, P > 0.05). In this condition, Per1 mRNA levels were persistently elevated (Fig. 2A; no significant difference with daytime levels in euthermic animals; P > 0.05), whereas those of Per2 were constantly depressed (Fig. 2B; no statistical difference with nighttime levels in euthermic animals; P > 0.05). For Bmal1, the nocturnal peak of expression was lost (Fig. 2C), and mRNA levels were intermediate between daytime and nighttime values of euthermic hamsters (P > 0.05 versus D and SD; P > 0.05 versus N). Altogether, these results show that the robust day/night oscillations of Per1, Per2, and Bmal1 expression disappear during hibernation, strongly suggesting that the circadian clockwork ceases to function. Rhythms with sluggish oscillations and extended periods seem unlikely because gene expression was not systematically restricted to intermediate levels. Importantly, we observed that the circadian molecular oscillations were clearly reexpressed during the interbout intervals (Fig. 2). This “recovery” and resynchronization of the clock were presumably facilitated by the occurrence of the light/dark cycle.
Fig. 2.
The circadian expression of clock genes in the SCN is abolished under deep hibernation. In situ hybridization was used to examine gene expression in euthermic and hibernating hamsters during daytime, nighttime, or during the subjective day. For all genes, the upper panel displays representative autoradiograms for each condition, and the lower panel shows the relative mRNA levels. (A–C) The expression of the clock genes Per1 (A), Per2 (B), and Bmal1 (C) exhibits circadian fluctuations in euthermic hamsters but not in hibernating animals. During hibernation, Per1 expression is elevated (A), Per2 expression remains low (B), and Bmal1 expression is intermediate between daytime and nighttime levels in euthermic hamsters (C). (D) Similarly, the expression of the clock-controlled gene Avp is rhythmic in euthermic but not in hibernating hamsters, in which its expression remains low. (E) The expression of the immediate early gene c-fos is dramatically increased in hibernating relative to euthermic hamsters, without significant day/night fluctuations. Data are shown as a percentage of the maximum and represent the means ± SEM (n = 4–7). Bars labeled with different characters (a, b, c) are significantly different from each other (P < 0.05).
Effect of Deep Torpor on Circadian Clock-Controlled Gene Expression.
If the molecular circadian clock is stopped during hibernation, then the 24-h rhythm of its output signals should be lost. Thus, we examined the expression of arginine vasopressin (Avp), a well characterized clock-controlled gene that constitutes a rhythmic output of the clock (6–8) and whose cyclic expression in the SCN depends directly on the molecular circadian machinery (25). As expected, Avp expression displayed a clear day/night change of expression in euthermic hamsters (Fig. 2D), with high mRNA levels during the day and the subjective day (P < 0.001, N versus D and SD). This change was in marked contrast with hibernating animals (P > 0.05), in which Avp expression was persistently depressed (Fig. 2D; for all conditions of hibernation, P > 0.05 versus euthermic N, and P < 0.001 versus euthermic D and SD). This result confirms and extends our previous conclusion, suggesting that the SCN molecular clockwork and at least one of its molecular outputs cease processing and transmitting circadian time.
Interestingly, the SCN maintains a relatively high metabolic activity during hibernation, as determined by analysis of [14C]2-deoxyglucose uptake and c-fos expression (26, 27). In the present work, examination of c-fos mRNA levels in the SCN also revealed a dramatic increase of expression in hibernating hamsters relative to euthermic animals (Fig. 2E; P < 0.001), without significant day/night differences (P > 0.05). This observation suggests that the loss of circadian organization in the SCN of hibernating hamsters does not simply result from reduced metabolic activity or transcription, and it favors the hypothesis that the SCN may serve a noncircadian role during hibernation (5).
SCN Output During Deep Hibernation.
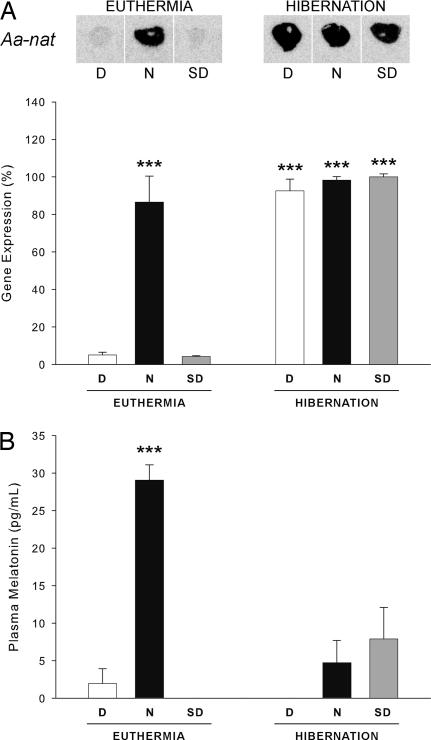
Finally, we questioned whether hibernation equally affects the rhythmic activity of SCN targets. The control of the rhythmic synthesis of melatonin by the pineal gland represents one of the best characterized outputs of the master clock (8). The SCN restricts production of melatonin to nighttime by controlling the activity of its rhythm-generating enzyme, arylalkylamine N-acetyltransferase (Aa-nat). In the European hamster, the daily variations of Aa-nat activity result from changes in transcription of the Aa-nat gene (28). Hence, the expression of this gene was used as an index of the pineal gland activation by the SCN. As expected in euthermic animals (28), Aa-nat expression was increased by >17-fold during nighttime (Fig. 3A; P < 0.001, N versus D and SD), which was faithfully translated into changes of plasma melatonin (P < 0.001, N versus D and SD), as measured by RIA (Fig. 3B). In contrast, Aa-nat mRNA levels were persistently high during hibernation (Fig. 3A) and were comparable with those of euthermic animals during nighttime (for all conditions of hibernation, P > 0.05 versus euthermic N, and P < 0.001 versus euthermic D and SD), suggesting that Aa-nat transcription is no longer rhythmic. This finding was confirmed at the melatonin level (Fig. 3B), although plasma melatonin concentrations were continuously low during torpor bouts (for all conditions of hibernation, P > 0.05 versus euthermic D and SD), as already described in other species (15–17). Interestingly, the plasma melatonin level has been reported to rise rapidly after arousal from hibernation, whatever the time of arousal (15, 16). The fact that Aa-nat mRNA levels remain persistently elevated during hibernation may well explain these observations and suggests a possible uncoupling between transcriptional activity and protein synthesis/enzymatic function.
Fig. 3.
Aa-nat is constitutively expressed in the pineal gland of hibernating hamsters. (A) Representative autoradiograms and quantification of Aa-nat mRNA demonstrate marked day/night changes of expression in euthermic animals, in contrast to hibernating hamsters, which display persistently high expression level. Data are shown as percentage of the maximum and represent the means ± SEM (n = 4–7). (B) Plasma melatonin concentration displays the expected large day/night difference in euthermic animals but not in hibernating hamsters, which exhibit constantly low levels. Data are means ± SEM (n = 4–7). ***, P < 0.001.
Discussion
Overall, our results show that the circadian clock of the SCN stops generating 24-h rhythms during hibernation, at least in the European hamster. We describe here the arrest of the master circadian clock over several days. The issue has been debated for many years (3, 5, 9), with various data either denying (10, 11, 13–17) or supporting (12, 18–20, 22) the existence of a functional pacemaker during deep torpor. Importantly, these contradictions may stem from species differences as well as from the variety of markers used to assess the functional state of the clock. Based on the recording of small circadian fluctuations in Tb, some reports have suggested the maintenance of circadian rhythms during hibernation (18–20). However, consistent with other reports (10, 14, 21), we did not detect such variations in the European hamster. In this work, we assessed gene transcription, thus we cannot rule out the possibility that the circadian clock might continue to function based on separate mechanisms, such as posttranscriptional regulation. Furthermore, the technique we used may not be suitable to detect either small subsets of cells that might retain temperature-compensated time measurement (20) or desynchronization of the individual SCN oscillators (9). However, we did not systematically observe intermediate mRNA levels, and the rhythmcity of both Aa-nat expression and plasma melatonin levels was lost during hibernation. The possibility that an extra-SCN oscillator could take over the circadian clock function is also unlikely because the SCN is one of the most active brain structures during hibernation (26). Furthermore, Hut et al. (10, 11) demonstrated that after the hibernation season, grounds squirrels display a period of arrhythmicity for both activity and Tb, supporting the absence of a circadian timer during hibernation. This arrhythmicity has been correlated with a loss of AVP immunostaining in the SCN during hibernation, which progressively reappears with circadian rhythmcity (11). Finally, Oklejewicz et al. (13) observed in Tau Syrian hamsters that mutation in the circadian system does not affect the torpor/arousal cycle (13). Curiously, it appears that the major difference between the studies that reported the persistence (12, 18–20, 22) or the absence (10, 11, 13–17) of a circadian signal during hibernation might be related to the Ta at which the experiments were conducted. This difference correlates with electrophysiological studies indicating that the circadian clock of hibernators still functions at temperatures lower than 37°C (5, 9), but recordings below 16.6°C failed to detect action potentials (29). Clearly, the disappearance of rhythmic oscillation in clock gene expression, together with the absence of action potentials, makes temperature compensation unlikely at low Tb (5–10°C). This property of the clock may be limited to a range of temperatures because rhythmic clock gene expression still occurs during daily torpor, a shallower form of torpor during which Tb decreases to 15–20°C for only several hours (30).
In conclusion, our data demonstrate that in the European hamster the molecular clock of the SCN is arrested during deep hibernation and no longer delivers circadian signals. Further experiments will be necessary to investigate the mechanism by which this arrest occurs. Examining the molecular clockwork during deep hibernation in other species and under different Ta will also reveal whether this phenomenon is species- and/or temperature-dependent.
Methods
Animals.
All experiments were performed in accordance with the rules of the French Department of Agriculture (license no. 67-38) and the European Committee Council Directive of November 24, 1986 (86/609/EEC). The adult male European hamsters used in this work were bred in house. The colony was established from animals caught in the field near Strasbourg, France, between 1994 and 1996. Hamsters were raised under seminatural conditions (natural photoperiod) with ad libitum access to water and food. In September, animals born in May of the previous year (16–17 months old) were transferred to a climatic room kept at 6 ± 2°C, under a 10-h light (150 lux; lights off at 1800)/14-h dark (2-lux dim red light) cycle. The core Tb was registered continually by thermosensitive radiotransmitters (model VM-FH-LT; Mini Mitter Co., Sunriver, OR) implanted in the abdominal cavity under isoflurane anesthesia, just before placing the hamsters in the climatic room. Radiofrequency signals from the implanted transmitters were averaged every 5 min by receivers placed under each animal's cage and collected by an automated computer software (Dataquest, St. Paul, MN). Straw was provided as nest-building material. All of the animals considered in this work had completed at least three bouts of torpor. Euthanasia occurred as depicted in Fig. 1C. Animals were anesthetized with N2O and killed by decapitation. Brains were rapidly removed from the skull, snap-frozen at −30°C, and stored at −80°C until in situ hybridization. Trunk blood was collected and centrifuged at 1,500 × g for 15 min, and plasma was stored at −20°C until melatonin assay.
In Situ Hybridization.
Radioactive in situ hybridization was performed as described previously (28), using riboprobes for rat Per1 (bases 638-1618 from GenBank accession no. NM_001034125), Per2 (bases 1170–1930 from NM_031678; Per1 and Per2 plasmids were kindly donated by H. Okamura, Department of Anatomy and Brain Science, Kobe University School of Medicine, Japan), Bmal1 (bases 75–1809 from AB012600), and Avp (bases 68–539 from M25646), or for Syrian hamster c-fos (bases 625-1025 from AF061881) and Aa-nat (bases 1–1045 from AF092100). After they were fixed, acetylated, and dehydrated, 20-μm-thick coronal brain sections were hybridized overnight at 54°C with 35S-UTP-labeled riboprobes. The sections were then treated with RNase A (10 μg/ml), washed (saline sodium citrate 0.1×, 60°C), dehydrated, and exposed to a Kodak BioMax MR Film (Sigma–Aldrich, Lyon, France) for 3–5 days along with 14C radioactive standards to allow standardization of densitometric measurements across films. X-ray films were scanned on an Epson 4990 transmittance scanner (Levallois-Perret, France), and background was subtracted. Calibrated optical density measurements of mRNA levels were performed by using ImageJ (National Institutes of Health, Bethesda, MD).
Melatonin RIA.
Plasma melatonin was analyzed by an RIA previously validated in European hamsters and described in ref. 28.
Data and Statistical Analyses.
Experiments were performed twice with similar results, and the data reported here are those from the second experiment. Results are shown as the percentage of the maximum and represent the mean ± SEM. Data were analyzed by two-way ANOVA, followed by HSD Tukey's analysis. Statistical significance was set at P < 0.05.
Acknowledgments
We thank Dr. David Hicks for English revision and Daniel Bonn for technical assistance. This work was supported by ACI Plates-formes d'exploration fonctionnelles thématisées from the French Ministry of Research and by the Centre National de la Recherche Scientifique and the Université L. Pasteur.
Abbreviations
- Aa-nat
arylalkylamine N-acetyltransferase
- Avp
arginine vasopressin
- Cry
cryptochrome
- D
daytime
- N
nighttime
- Per1
period 1
- Per2
period 2
- SCN
suprachiasmatic nuclei of the hypothalamus
- SD
subjective day
- Ta
ambient temperature
- Tb
body temperature.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Geiser F. Annu Rev Physiol. 2004;66:239–274. doi: 10.1146/annurev.physiol.66.032102.115105. [DOI] [PubMed] [Google Scholar]
- 2.Carey HV, Andrews MT, Martin SL. Physiol Rev. 2003;83:1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- 3.Kortner G, Geiser F. Chronobiol Int. 2000;17:103–128. doi: 10.1081/cbi-100101036. [DOI] [PubMed] [Google Scholar]
- 4.Kilduff TS, Krilowicz B, Milsom WK, Trachsel L, Wang LC. Sleep. 1993;16:372–386. doi: 10.1093/sleep/16.4.372. [DOI] [PubMed] [Google Scholar]
- 5.Ruby NF. J Biol Rhythms. 2003;18:275–286. doi: 10.1177/0748730403254971. [DOI] [PubMed] [Google Scholar]
- 6.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hastings MH, Herzog ED. J Biol Rhythms. 2004;19:400–413. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- 8.Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- 9.Heller HC, Ruby NF. Annu Rev Physiol. 2004;66:275–289. doi: 10.1146/annurev.physiol.66.032102.115313. [DOI] [PubMed] [Google Scholar]
- 10.Hut RA, Barnes BM, Daan S. J Comp Physiol B. 2002;172:47–58. doi: 10.1007/s003600100226. [DOI] [PubMed] [Google Scholar]
- 11.Hut RA, Van der Zee EA, Jansen K, Gerkema MP, Daan S. J Comp Physiol B. 2002;172:59–70. doi: 10.1007/s003600100227. [DOI] [PubMed] [Google Scholar]
- 12.Daan S. Neth J Zool. 1973;23:237–265. [Google Scholar]
- 13.Oklejewicz M, Daan S, Strijkstra AM. J Comp Physiol B. 2001;171:431–439. doi: 10.1007/s003600100193. [DOI] [PubMed] [Google Scholar]
- 14.Wollnik F, Schmidt B. J Comp Physiol B. 1995;165:171–182. doi: 10.1007/BF00260808. [DOI] [PubMed] [Google Scholar]
- 15.Vanecek J, Jansky L, Illnerova H, Hoffmann K. Comp Biochem Physiol A Physiol. 1985;80:21–23. doi: 10.1016/0300-9629(85)90671-1. [DOI] [PubMed] [Google Scholar]
- 16.Stanton TL, Craft CM, Reiter RJ. J Exp Zool. 1986;239:247–254. doi: 10.1002/jez.1402390212. [DOI] [PubMed] [Google Scholar]
- 17.Florant GL, Rivera ML, Lawrence AK, Tamarkin L. Am J Physiol. 1984;247:R1062–R1066. doi: 10.1152/ajpregu.1984.247.6.R1062. [DOI] [PubMed] [Google Scholar]
- 18.Pohl H. Z Vergl Physiol. 1961;45:109–153. [Google Scholar]
- 19.Menaker M. Nature. 1959;184:1251–1252. [Google Scholar]
- 20.Grahn DA, Miller JD, Houng VS, Heller HC. Am J Physiol. 1994;266:R1251–R1258. doi: 10.1152/ajpregu.1994.266.4.R1251. [DOI] [PubMed] [Google Scholar]
- 21.Canguilhem B, Malan A, Masson-Pévet M, Nobelis P, Kirsch R, Pévet P, Le Minor J. J Comp Physiol B. 1994;163:690–698. doi: 10.1007/BF00369521. [DOI] [PubMed] [Google Scholar]
- 22.French AR. J Comp Physiol B. 1977;115:87–100. [Google Scholar]
- 23.Hermes ML, Buijs RM, Masson-Pévet M, van der Woude TP, Pévet P, Brenkle R, Kirsch R. Proc Natl Acad Sci USA. 1989;86:6408–6411. [Google Scholar]
- 24.Magarinos AM, McEwen BS, Saboureau M, Pévet P. Proc Natl Acad Sci USA. 2006;103:18775–18780. doi: 10.1073/pnas.0608785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 26.Kilduff TS, Sharp FR, Heller HC. J Neurosci. 1982;2:143–157. doi: 10.1523/JNEUROSCI.02-02-00143.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bitting L, Sutin EL, Watson FL, Leard LE, O'Hara BF, Heller HC, Kilduff TS. Neurosci Lett. 1994;165:117–121. doi: 10.1016/0304-3940(94)90723-4. [DOI] [PubMed] [Google Scholar]
- 28.Garidou ML, Vivien-Roels B, Pévet P, Miguez J, Simonneaux V. Am J Physiol. 2003;284:R1043–R1052. doi: 10.1152/ajpregu.00457.2002. [DOI] [PubMed] [Google Scholar]
- 29.Miller JD, Cao VH, Heller HC. Am J Physiol. 1994;266:R1259–R1266. doi: 10.1152/ajpregu.1994.266.4.R1259. [DOI] [PubMed] [Google Scholar]
- 30.Herwig A, Revel F, Saboureau M, Pévet P, Steinlechner S. Chronobiol Int. 2006;23:269–276. doi: 10.1080/07420520500522424. [DOI] [PubMed] [Google Scholar]