Abstract
In this study, a quantitative approach was used to investigate the role of D142, which belongs to the highly conserved E/DRY sequence, in the activation process of the α1B-adrenergic receptor (α1B-AR). Experimental and computer-simulated mutagenesis were performed by substituting all possible natural amino acids at the D142 site. The resulting congeneric set of proteins together with the finding that all the receptor mutants show various levels of constitutive (agonist-independent) activity enabled us to quantitatively analyze the relationships between structural/dynamic features and the extent of constitutive activity. Our results suggest that the hydrophobic/hydrophilic character of D142, which could be regulated by protonation/deprotonation of this residue, is an important modulator of the transition between the inactive (R) and active (R*) state of the α1B-AR. Our study represents an example of quantitative structure-activity relationship analysis of the activation process of a G protein-coupled receptor.
The α1B-adrenergic receptor (AR) belongs to the superfamily of G protein-coupled receptors (GPCRs). The seven transmembrane domains (TMDs) common to all GPCRs contribute to the formation of the ligand binding pocket, whereas aa sequences of the intracellular loops (i) appear to mediate receptor–G protein coupling (1, 2). However, how binding of the extracellular signals is converted into receptor activation remains largely unknown.
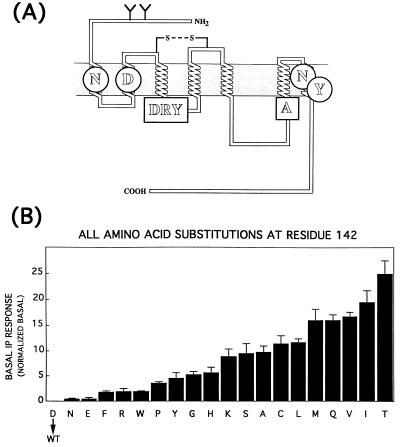
Recently, we have investigated the activation process of the α1B-AR linked to phospholipase C-mediated activation of polyphosphoinositide hydrolysis. By a combination of site-directed mutagenesis of the α1B-AR with computational simulations of receptor dynamics we explored the potential molecular mechanisms underlying the process of receptor activation, by focusing on a number of constitutively active receptor mutants (3). We identified a series of molecular changes that appear to be correlated with the transition from the inactive (R) to the active (R*) state, independently of the presence of the agonist. We proposed that the equilibrium between R and R* of the α1B-AR depends, at least in part, on the prototropic equilibrium between the deprotonated (anionic) and protonated (neutral) forms of D142, the negatively charged residue present in the E/DRY motif, which is highly conserved among different GPCRs (Fig. 1A). As a result, we found that replacement of D142 with the nonpolar aa alanine conferred high constitutive (agonist-independent) activity to the α1B-AR. According to our analysis, a series of intramolecular interactions that might be of fundamental importance in the process of receptor activation depends on the protonation state of D142. In particular, our model pointed to a conserved “polar pocket” formed near the cytosol via a network of H-bonding interactions among N63, D91, N344, and Y348 (Fig. 1A). This set of interactions constrains the receptor in its inactive state by exerting control on the degree of cytosolic exposure of the arginine residue located in the E/DRY sequence, in a manner conditionally coupled to proton transfer onto D142. When this aspartate is in its anionic form, the interaction of R143 with D91 is highly favored, and this maintains the arginine residue and several other amino acids of the second (i2) and third (i3) intracellular loops in a “buried” condition with respect to the cytosol. In contrast, the active states of the receptor, induced either by protonation of D142 (R*) or by constitutively activating mutations, although not structurally identical, all show R143 shifted out of the polar pocket.
Figure 1.
(A) Topographical model of α1B-AR. The figure highlights the amino acids N63, D91, N344, and Y348 forming the polar pocket (circles), the DRY sequence, and A293 (squares). (B) Constitutive activation of α1B-AR by all aa substitutions at position 142. Basal levels of IP accumulation were measured in COS-7 cells expressing the wild-type α1B-AR (WT) or mutant receptors carrying different amino acids at position 142. The basal IP response was calculated as indicated in the legend of Table 1. The results are from several independent experiments, each performed in triplicate.
However, we also noted that although the replacement of D142 with a hydrophobic residue (D142A) resulted in very high levels of constitutive activity, the substitution with residues of either neutral (D142N) or positive charge (D142R) conferred only very modest agonist-independent activity to the receptor. Thus, the mere suppression of a negative charge cannot be the only driving force that triggers constitutive activity, suggesting that other physicochemical properties of the residue in position 142 must be implicated.
To better understand how the nature of the residue in such a position influences the transition from inactive to active conformations, we performed experimental and computer-simulated mutagenesis by substituting all possible natural amino acids at the D142 site of the α1B-AR. The resulting congeneric set of proteins–i.e., differing for only a single residue in the same position–together with the finding that all the receptor mutants displayed various levels of constitutive activity enabled us to quantitatively analyze the relationships between their structural/dynamic features and the extent of agonist-independent activity. To our knowledge, this study represents the first successful attempt to apply a quantitative structure-activity relationship analysis to the description of the intrinsic process of the activation of GPCRs.
MATERIALS AND METHODS
Construction of the α1B-AR Mutants and Cell Transfection.
The site-specific PCR-mediated mutagenesis and the DNA transfection in COS-7 cells were performed as described (4). The cDNA encoding different receptors was subcloned in the expression vector pRK5.
Ligand Binding and Inositol Phosphate Determination.
Membrane preparations derived from cells expressing the different α1B-ARs, ligand binding experiments using [125I]HEAT (DuPont/NEN), and the determination of labeled inositol phosphates in cells labeled with [3H]inositol (Anawa, Zurich) (5–10 μCi/ml for 15–18 hr; 1 Ci = 37 GBq) were also performed as described (4, 5).
Computer-Simulated Mutagenesis.
Three-dimensional model building and molecular dynamics simulations of the α1B-AR mutant structures were performed according to the computational procedure extensively described in our previous work (3).
Modeling studies as well as solvent accessible surface area calculations (6) were performed with the molecular graphics package quanta (version 4.0; Molecular Simulations). Energy minimizations and molecular dynamics simulations were done using the program charmm (version 22; ref. 7).
RESULTS
Constitutive Activity of the α1B-AR Carrying Amino Acid Substitutions at Position 142.
D142 of the α1B-AR belonging to the highly conserved E/DRY motif was mutated into all 19 possible natural amino acids, and the receptor mutants were tested for their ability to induce constitutive (agonist-independent) inositol phosphate (IP) accumulation in transfected COS-7 cells. All 19 substitutions of D142 resulted in α1B-AR mutants displaying different levels of constitutive activity ranging from 18 to 698% above control levels of the mock-transfected cells (Table 1). The constitutive activity of all 19 mutants was inhibited by the α1B-AR antagonist prazosin (results not shown). On the other hand, in this series of experiments the agonist-independent IP accumulation in COS-7 cells expressing the wild-type α1B-AR was similar to that of mock-transfected cells.
Table 1.
Ligand binding properties and IP accumulation in COS-7 cells expressing wild-type αIB-AR or mutant receptors carrying different amino acids at position 142.
| [125I] HEAT binding
|
3H-IP accumulation
|
||||
|---|---|---|---|---|---|
| Receptor | Bmax, pmol/mg | EPI, Ki (μM) | Rmax, % increase | Basal IP, % control | Normalized basal |
| αIB-AR (9) | 1.17 | 12.80 | 375 ± 55 | 0 | 0.00 |
| D142N (6) | 0.55 | 9.42 | 320 ± 79 | 23 ± 11 | 0.42 |
| D142E (6) | 0.78 | 11.30 | 297 ± 27 | 39 ± 15 | 0.50 |
| D142F (4) | 0.47 | 3.45 | 200 ± 71 | 82 ± 14 | 1.74 |
| D142R (6) | 0.14 | 0.24 | 50 ± 14 | 27 ± 9 | 1.93 |
| D142W (4) | 0.25 | 1.81 | 30 ± 1 | 49 ± 2 | 1.96 |
| D142P (4) | 0.05 | 3.50 | 59 ± 13 | 18 ± 1 | 3.60 |
| D142Y (5) | 0.23 | 2.73 | 41 ± 14 | 104 ± 26 | 4.52 |
| D142G (5) | 0.24 | 3.20 | 144 ± 33 | 126 ± 14 | 5.25 |
| D142H (4) | 0.13 | 1.11 | 66 ± 21 | 73 ± 14 | 5.62 |
| D142K (4) | 0.13 | 0.30 | 22 ± 16 | 115 ± 19 | 8.85 |
| D142S (3) | 0.38 | 0.55 | 25 ± 16 | 361 ± 70 | 9.50 |
| D142A (3) | 0.34 | 0.45 | 36 ± 16 | 329 ± 42 | 9.68 |
| D142C (5) | 0.37 | 0.73 | 13 ± 8 | 422 ± 58 | 11.41 |
| D142L (4) | 0.35 | 1.10 | 48 ± 27 | 407 ± 26 | 11.63 |
| D142M (4) | 0.19 | 1.70 | 57 ± 21 | 302 ± 41 | 15.89 |
| D142Q (4) | 0.37 | 0.37 | 38 ± 24 | 599 ± 43 | 15.89 |
| D142V (5) | 0.23 | 0.37 | 14 ± 4 | 382 ± 21 | 16.61 |
| D142I (3) | 0.22 | 0.52 | 8 ± 1 | 427 ± 49 | 19.41 |
| D142T (5) | 0.28 | 0.80 | 10 ± 4 | 698 ± 74 | 24.93 |
The amount of transfected DNA/106 cells was calibrated for each receptor mutant to achieve expression levels ranging 0.7–0.2 pmol/mg of protein (0.17–0.05 pmol/106 cells) for most receptor mutants. [125I]HEAT and epinephrine (EPI) binding as well as IP accumulation were measured as described. Rmax indicates the EPI-induced IP accumulation expressed as percentage increase above basal levels in the absence of EPI. Basal IP indicates the IP levels in the absence of EPI expressed as percentage increase above those of mock-transfected cells (control). Normalized basal indicates the ratio between the basal IP values divided by 100 and receptor number (pmol/mg of protein). Results are the mean ± SE of several independent experiments whose numbers are indicated in parentheses. The Ki values were from two to three experiments that agreed within 20%.
Because most of the mutant receptors did not express to the same extent as the wild-type receptor did in COS-7 cells, we studied the relationship between expressed receptors and extent of constitutive activity for a majority of highly active mutants. By manipulating the concentration of transfected DNA, we obtained a sufficiently wide range of membrane expression for each receptor examined.
Within the range, the constitutive activity of the receptor mutants was always linearly dependent on receptor density and did not show any trend for saturation (results not shown). Thus, the extent of constitutive activity was expressed as the ratio between basal IP and receptor expression (Normalized basal; Table 1). Both these quantities were routinely measured in parallel in each experiment. It must be noted, however, that the error inherent in this transformation is correspondingly larger for those mutants that did not achieve expression levels greater than 0.15 pmol/mg of proteins, such as those carrying basic aa substitutions (R, K, and H) or proline (P) (Table 1).
As shown in Fig. 1B, there is a smooth relationship between the extent of constitutive activity and the type of residue replaced at position 142, which is qualitatively different from that previously described for the analogous set of substitutions made at position 293 of the α1B-AR (4). It is also clear that the basal responses (Table 1) observed for the most active mutants were almost 2-fold higher than the maximal response induced by the agonist (Rmax; Table 1) on the wild-type receptor. Moreover, all the studied mutants, including those with the highest level of constitutive activity, maintained the ability to respond to the agonist. Thus, an important conclusion that can be drawn from these data is that several of the mutants generated in this study are not simply constitutively active—they are hyperactive, in the sense that their intrinsic ability to activate the G protein is even greater than that of the wild-type receptor in the agonist-bound form.
Binding isotherms of the antagonist prazosin and the agonist epinephrine in competition for the binding sites labeled by [125I]HEAT were obtained for all 19 mutants. We did not observe substantial differences in antagonist-binding affinity between the wild-type α1B-AR and any of the 19 mutants (data not shown). In contrast, there were major changes in agonist affinity. In fact, with the exception of D142N and D142E, all receptor mutants displayed from 4- to 40-fold smaller Ki for epinephrine compared with the wild-type α1B-AR (Table 1). Because the residue in position 142 cannot provide direct interactions with the molecule of the ligand, it is clear that such agonist-selective changes of affinity are cooperative in nature and reflect the conformational shifts toward the active state(s) that each substituted residue imposes to a different extent in the receptor. As shown in Table 1, the enhancement of agonist affinity was essentially proportional to the extent of constitutive activity for the majority of receptor mutants. Notable exceptions were the two cationic residue mutations, D142R and D142K, which enhanced affinity much more than basal activity, and, conversely, D142M and D142T, which produced a very high level of constitutive activity with a proportionally smaller increase of binding affinity.
Quantitative Relationship Between the Hydrophobic Character of the Replaced Amino Acids and Constitutive Activity.
The challenging finding that all 19 possible aa substitutions of D142 induce graded levels of constitutive activity prompted us to search for quantitative relationships between empirical descriptors of the physicochemical properties of the natural amino acids and the degree of the agonist-independent activity of the α1B-AR mutants.
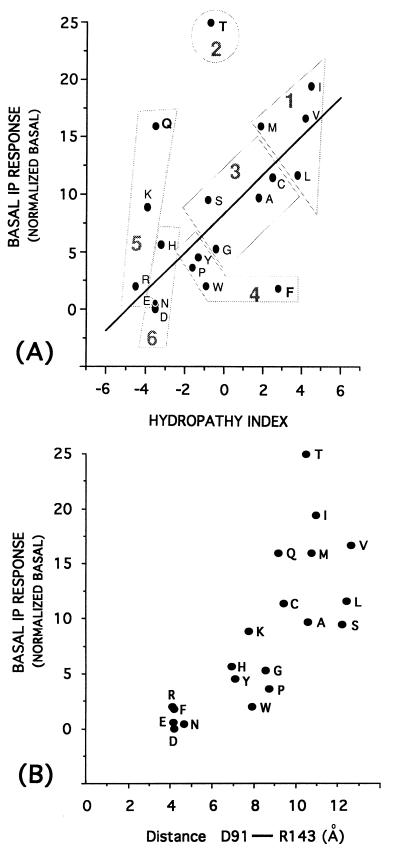
We analyzed a total of 96 descriptors of the physicochemical properties of natural amino acids, including a variety of hydrophobicity and hydrophilicity parameters (8–10), size descriptors (11), volume and surface area values (8, 12–14), solution properties (8), chromatographic properties (8), R values (8, 15), polarity and polarizability indices (8, 12), and the average electrostatic field computed on the van der Waals surface of the residue side chains (16). The simple regression analysis revealed that the most significant linear trends with the constitutive activity values are reached by several hydrophobicity parameters (17–20), which are intercorrelated. Among them, those proposed by Kyte and Doolittle (17) for natural amino acids and based on both water-vapor transfer energies and the interior-exterior distribution of the side chains (21) yielded the best trend (i.e., the lesser number of outliers within all the 20 natural aa determinations) against the degree of constitutive activity (Fig. 2A). The correlation becomes even tighter if we exclude from the analysis three residues (T, F, and Q) that are evident outliers. In this case, up to 70% of the overall variation in basal receptor response is explained by the empirical descriptor, which indicates that the extent of constitutive activity in the receptor is increased by 7–8% for each unit of enhancement in hydrophobic character of the residue in position 142. Hydrophobicity is thus the most important quantitative descriptor of the extent to which a mutation at this site is likely to promote the conversion of the receptor into its active form.
Figure 2.
(A) Correlation between the constitutive activity (basal IP response) of the 19 mutants of D142 and the hydropathy indices of the replaced amino acids. Linear regression equation: Basal IP response = 8.29 (± 0.81) + 1.69 (± 0.27) × hydropathy index; n = 17, r = 0.85, s = 3.2, where n is the number of compounds, r is the correlation coefficient, s is the standard deviation, and the numbers in parentheses are the 95% confidence intervals. The three outliers, F, Q, and T, have been discarded from correlation. The figure also shows the six groups in which the amino acids can be clustered based on the principal component analysis previously described considering 29 of their different experimental properties (22). This analysis identifies three main components, z1, z2, and z3, whose values are regarded as principal properties of the natural amino acids. z1 is mainly related to hydrophilicity, z2 is additionally influenced by size and some hydrophobicity/hydrophilicity scales, and z3 contains information from the analysis of pKa, pI, and 1H-NMR values. (B) Correlation between the constitutive activity (basal IP response) of the 19 mutants of the D142 site and the distance between the γ-C-atom of D91 (TMDII) and the ζ-C-atom of R143 (TMDIII). This distance was measured on the averaged minimized structures of the α1B-AR wild type carrying D142 deprotonated and of the 19 mutants of D142.
However, because the outliers identified in this analysis are not poorly expressed mutants (the activity of which is likely to be biased by a larger experimental error) and belong to the amino acids showing the most unequivocal hydropathy values (17), additional properties of the aa replaced in position 142 must also contribute to modulate the constitutive activities of the receptor mutants.
A recent application (22) of the principal component analysis to 29 different experimental properties of the 20 coded amino acids identified three principal components mainly related, respectively, to hydrophilicity (z1), bulk (z2), and electronic (z3) properties. According to these components, amino acids are clustered into six groups of gradually decreasing similarity (going from group 1 to 6 in Fig. 2A). Interestingly, as shown in Fig. 2A, there is a broad correspondence between the rank order of group similarity and the degree of constitutive activity induced by the mutated residue. Thus, the principal components may help elucidate the behavior of the threonine and phenylalanine, which are outliers in the correlation between the hydropathy indices and constitutive activity values. In fact, the major outlier, threonine, is classified as a singular aa in principal component analysis, because its size is close to that of members of group 1, but its hydrophilic properties are typical of the more polar members of group 3. Because this residue caused similar or even greater constitutive activity than residues of group 1, its size and shape features may influence the constitutive activity more than its hydrophilic properties. Analogous considerations apply to phenylalanine, which shares the activating properties of other aromatic residues in group 4 but exhibits the hydrophobic character distinctive of group 1. Yet, not even principal component analysis ranking can explain the behavior of glutamine, the third outlier (Fig. 2A). Additional mechanisms, including, perhaps, direct interaction with regulatory proteins, might underlie the constitutive activity of the D142Q mutant.
The finding that a local molecular property, such as the hydrophobic character of a single residue, is so strongly linked to the global conformational change underlying the conversion of a macromolecule like the α1B-AR into an active form has very important mechanistic implications. This is particularly true in our context, in which the mutated residue is a highly conserved aspartate whose hydrophobic character can change reversibly depending on its protonation state. In fact, it has been shown that the hydrophobicity parameters of the anionic form of aspartate or glutamate are similar to those of the highest hydrophilic residues, whereas their neutral (protonated) forms have values comparable to the hydrophobicity of alanine (23). Thus, the protonation of D142, by increasing the hydrophobicity of this residue at comparable levels as that of alanine, might trigger wild-type receptor activation similar to the constitutively active D142A mutant. According to the meaning of the hydrophobicity as empirical descriptor (obtained as mean values of the free energy transfer from a polar to a nonpolar environment), the mechanism triggering receptor activation may consist in the ability of D142 to translocate from the cytosolic water to a less polar environment. To gain a better understanding of the structural effects of mutation as well as of protonation of D142, computer-simulated mutagenesis was thus undertaken on the three-dimensional model of α1B-AR.
A Theoretical Descriptor of Motion in the DRY Motif Predicts Constitutive Activity.
To produce computer-generated mutant structures, we simulated all the possible 19 aa substitutions at position 142 of the α1B-AR and compared and minimized the structures averaged over the last 100 ps of the molecular dynamics simulations. The theoretical models obtained allowed us to compute both theoretical descriptors localized on the mutation site (D142) as well as descriptors related to the structural/dynamic effect of the D142 substitutions.
Because the local descriptor was most closely related to the hydropathy index, we computed the solvent accessible surface area (6) of the amino acid 142. Since this measurement yielded zero values for many residues, we could not calculate a reliable coefficient for the correlation between basal activity and magnitude of the solvent accessible surface area. Yet, the negative relationship between these two variables is straightforward. In fact, all residues causing the largest level of constitutive activation (S, A, L, C, M, V, I, and T) were found completely shielded from the solvent, very small accessible areas were measured for those inducing intermediate levels of activity (P, Y, G, and K), whereas the largest exposed areas were associated with residues causing small or no levels of activation. Again, the D142Q mutant exhibited a glaring deviation from this relationship because its solvent-exposed surface values were close to those of weakly activating mutants, despite its relatively high constitutive activity.
The correspondence between hydrophobicity, reduction of solvent accessible surface area, and constitutive activity suggests a precise mechanism for mutationally induced activation of the α1B-AR: the side-chain of the residue replaced in position 142 has to become buried to allow the interconversion of the molecule into an active form.
Interesting findings result from the comparison of the two average minimized structures of the wild-type α1B-AR carrying D142 in both its prototropic forms. In fact, the side chain of D142 in the average structure of the wild-type receptor carrying this residue in the anionic form shows 39 Å2 of surface available to the solvent, whereas this value drops to zero in the structure carrying D142 in its protonated form. Thus, protonation of D142 appears to be a sufficient change to mimic the effect of mutations with hydrophobic residues, suggesting that burying its side chain may be the fundamental molecular step common to the active forms of both wild-type and mutant receptors.
Identification of a Structural/Dynamic Feature Related to the Burying of the Side Chain in Position 142.
We have previously shown that a major common structural feature shared by wild-type α1B-AR carrying D142 in its protonated form and high constitutively active mutants is the cytosolic exposure of R143 (3). Here, through a detailed comparison of the structures of the 19 mutants with those of the wild-type receptor in both prototropic forms, we addressed the question of whether the hydrophobically mediated internalization of residue 142 may be mechanistically connected to the release of R143 from the polar pocket, which marks the transition of the receptor to its active form.
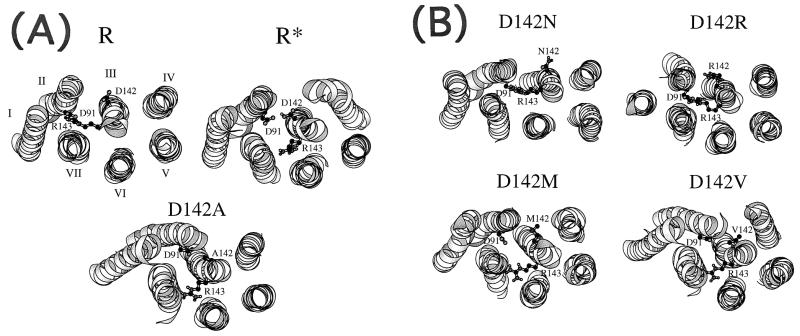
We identified a series of molecular changes that appear to link the motion of these two residues in an opposite fashion. First, we found that when D142 is in its anionic form, its side chain is directed toward the cytosolic “water” and it is stabilized by a direct salt bridge with R160 of the i2. Although such interaction may be overemphasized in our structures, which are results of simulations performed in vacuo, it is conceivable that the attraction between the two charged residues still operates in the solvated protein, perhaps through a network of shared water molecules. Protonation of D142, or its replacement by a hydrophobic residue, invariably breaks the coulombic interaction with R160 and triggers internalization of the residue into a water-shielded environment, where van der Waals interactions with the surrounding amino acids of the receptor may be a major stabilizing factor. The pattern of these interactions depends on the chemical nature of the aa replaced at position 142. Second, the transit of residue 142 to a more buried position appears to set for the larger scale movements that propagate through the whole transmembrane bundle. Rigid body movements of TMD III, consisting in rotations, translations, and vertical displacements toward the cytosol, result in the internalization of its cytoplasmic end (Fig. 3). Other helices and their connected intracellular loops also undergo extensive changes, including the kink imposed by proline 345 to the main axis of TMD VII.
Figure 3.
(A) View of the minimized average structures of wild-type α1B-AR (R and R*) and of the D142A mutant in a direction parallel to the helix main axes from the intracellular side. The R structure carries D142 in the deprotonated (anionic) form, whereas the R* structure carries D142 in the protonated (neutral) form. For clarity, only the α-helical domains have been represented, whereas the loops have been excluded from the picture. (B) View of the minimized average structures of the D142N, D142R, D142M, and D142V mutants in a direction parallel to the helix main axes from the intracellular side.
The important consequence of this concerted set of modifications is that R143 of the DRY motif is progressively pulled away from D91, which is part of the polar pocket resting between TMD VI and VII (Fig. 3). If the pulling of R143 toward the cytosol is the principal consequence of the structural changes induced by the modifications at the D142 site, we should expect that a descriptor able to quantify the gradual release of R143 observed in the 19 mutant structures should also be correlated with experimentally measured constitutive activity. Thus, we measured the distance between the γ-C-atom of D91 and the ζ-C-atom of R143 in the average minimized structures of the wild-type as well as of all 19 mutated α1B-AR. As shown in Fig. 2B, we found a clear-cut relationship between the extent of the shift of R143 and the degree of agonist-independent activity of the receptor mutants. The higher the extent of constitutive activity of the mutant, the larger the distance between D91 and R143 in the average minimized mutated structures (Fig. 2B). In the wild-type α1B-AR, the D91-R143 distance used in this correlation (4.2 Å) was measured on the average structures carrying D142 in its deprotonated form. Interestingly, in the wild-type α1B-AR carrying the protonated D142, the same measured descriptor is 9.63 Å. Thus, the protonation of D142 is predicted to lead to an active form of the α1B-AR.
DISCUSSION
We have studied a new series of constitutively active mutants of α1B-AR generated by replacing the aspartic residue in position 142 with all other natural amino acids. As in a previous study, where all-residue scanning mutagenesis was used to target alanine 293 in the C-terminal end of i3, the variation of constitutive activity can be univocally related to the identity of the point-mutated residue. However, important differences characterize this new study. First, the targeted residue is part of the conserved DRY sequence (Fig. 1A), which is shared by a majority of members of the 7TM families and plays a key role in the process of receptor activation, as both molecular modeling and experimental evidence indicate (24–27). Second, there is a quantitative relationship between the empirically deduced hydropathy index of the substituted residues and the extent of agonist-independent activity. The important implication of this quantitative relationship is that the same physicochemical property that modulates the activities of the receptor mutants might also regulate the reversible transition of the wild-type α1B-AR from its inactive to active form(s), by means of a prototropic equilibrium between deprotonated and protonated forms, respectively, of the targeted residue D142.
Our results provide consistent explanations of a number of experimental observations whose common mechanistic link had remained obscure thus far. One is the intriguing role that residues D142 and A293 appear to play in maintaining the α1B-AR in the inactive form. Both exhibit a compelling intolerance to mutation, because virtually any replacement with other residues causes receptor activation to some extent. Yet, they are located on diverse receptor domains and mark the cytosolic poles of two distinct transmembrane helices. We have shown that the relationship between the type of mutated residue and the extent of constitutive activity is completely different for the two series of mutants, which implies a difference in the way they trigger activation. In our models (3), A293 lies on the C-terminal portion of i3 with its side chain oriented toward several residues of the α-helical N-terminal portion of that loop. Any alternative side chain introduced by mutation changes the interhelical interaction pattern and consequently promotes, through a complex series of intramolecular interactions, rigid helix movements of TMD III, culminating in the shift of R143 away from the polar pocket. In contrast, as described in this study, D142 must be negatively charged and well exposed to the cytosol to maintain the inactive conformation. Any side chain replacement increasing the hydrophobicity of residue 142 results in sufficient internalization of this residue and can directly propel the motion of TMD III. Thus, different mechanisms trigger constitutive activation at the 293 and 142 sites: a gain of any kind of interaction that indirectly affects TMD III operates in the first; a decrease of electrostatic as well as H-bonding interactions directly influencing the position of TMD III accounts for the second. The final outcome, however, is identical and consists in the shift of R143 out of the polar pocket (Fig. 3). Accordingly, an interatomic distance between R143 and a residue of the polar pocket can be a quantitative predictor of the extent of experimentally measured constitutive activity (Fig. 2). On the basis of this comparative study, we propose that the main role of A293 is to constrain the α1B-AR in its inactive state, and that a similar role can be played by other residues lying in different interhelical positions.
Another important experimental observation demanding explanation is the connection found in rhodopsin between proton uptake by the anionic residue of the E/DRY sequence and receptor activation (28). We have previously described substantial structural similarity between the minimized average structure of the wild-type α1B-AR carrying D142 in its protonated form and those of constitutively active mutants (3). On this basis, we proposed that protonation of D142 may be one of the molecular steps conditionally dependent on the agonist-induced transition of a receptor into an active form. However, the underlying mechanism linking protonation of D142 to receptor activation remained unclear. Here, we suggest that the change of hydrophobicity of D142 upon its protonation promotes internalization of its side chain, which is a sufficient thrust for motion in TMD III to occur. This is supported by the high correlation between the hydropathy index of the substituted amino acids at position 142 and the constitutive activity of the 19 mutants.
Although this study was primarily dedicated to elucidate mutationally induced activation of the α1B-AR, the proposed mechanism of proton-mediated regulation of a single residue might provide new clues in understanding how agonists activate wild-type GPCRs. In fact, an important prediction of this study is that the conformational change of the α1B-AR induced by an agonist could be coupled to a change of the pKa of D142 by changing its environment and thereby shifting its prototropic equilibrium toward protonation. Experimental support of this hypothesis is found in the work of Cohen et al. (26), who studied the constitutive activation induced in rhodopsin by the mutagenesis of K296, the site of attachment of retinal to the protein through a protonated Schiff-base linkage. From the bell-shaped pH-dependence of the receptor-mediated GTPase activity of transducin for rhodopsin mutants, an apparent pKa2 value was computed that described the propensity of rhodopsin to enter the active state, and it was inversely correlated to the volume of the replaced residue. As E134 of the E/DRY motif (corresponding to D142 in the α1B-AR) seems to be one of the main amino acids contributing to pKa2, Cohen’s findings strongly suggest that proton binding on a potential G protein docking region of rhodopsin is allosterically linked to conformational changes in the retinal binding site (26).
If we take the structure of wild-type α1B-AR with protonated D142 as the closest equivalent in our study of the agonist-induced R* conformation, two interesting considerations follow. First, the hydrophobicity index of the protonated D142 (23) and the extent of the R143 shift measured in the average structure of the wild-type carrying D142 protonated (9.63 Å) are both smaller than the corresponding values associated with mutants displaying very high levels of constitutive activity. Therefore, according to the magnitude of the conformational changes, those mutants are hyperactive rather than only constitutively active. Similar conclusions were drawn from experimental data when comparing the extent of constitutive activity of the mutants with the maximal agonist-induced stimulation of the wild-type receptor (Table 1). Second, many of the hyperactive mutants carrying hyrophobic residues in position 142 still respond to the agonist with a further increase of IP response. This suggests that the mechanism through which the agonist might affect the pKa of D142 may also be essentially hydrophobic in nature, such as, for example, a conformationally induced crowding of nonpolar side chains in the local environment of this residue. In this case, a highly hydrophobic residue can undergo further internalization and induce additional strain to TMD III and, thus, further stabilization of the active conformations.
In conclusion, we propose that D142 plays a crucial role in regulating the reversible transition of the α1B-AR from R to R* through protonation events of its side chain. We also suggest that the agonist-dependent switch in this equilibrium crucially involves the pKa of D142. Electrostatic and hydrophobic interaction-induced pKa shifts of titratable amino acids often have been implicated in crucial steps of enzyme catalysis and protein functions including the photooscillating proton transport in bacteriorhodopsin (29, 30).
We have observed that the main structural effect of the burying of the replaced residue at the D142 site as well as of the protonated side chain of D142 is the shift of R143 out of the polar pocket, which promotes the exposition of several cationic amino acids on the C-terminal portion of i3 (i.e., R288 and K291) toward the cytosol. This translocation of positively charged amino acids could represent a fundamental driving force in the process of receptor–G protein recognition, as it is well known that long-range-acting electrostatic forces play a fundamental role in protein–protein recognition. Moreover, the rearrangement of charged as well as hydrophobic amino acids following mutation or agonist-induced protonation of D142 could promote further pKa shifts involving other titratable amino acids in the G protein binding domain of the receptor and culminating into proton release to the G protein.
Altogether, the implications of our study are twofold. First, it provides a mechanistic hypothesis about the role played by the highly conserved E/DRY motif in the activation process of GPCRs. Second, it provides evidence that a quantitative structure-activity relationship analysis can help elucidate the intrinsic processes underlying the function of this class of macromolecules.
Acknowledgments
We thank Nadia Larbi for her technical assistance in the site-directed mutagenesis work. This work was supported by the European Community (Biotechnology Project BIO2CT-930083-EUROCEPTOR) to T.C. and S.C., a grant from the Fonds National Suisse de la Recherche Scientifique (31–33684.92) to S.C., and the Consiglio Nazionale delle Ricerche and the Ministero dell’ Universitá e della Ricerca Scientifica e Tecnologica of Italy (40% funds) to P.G.D.
Footnotes
Abbreviations: α1B-AR, α1B-adrenergic receptor; GPCR, G protein-coupled receptor; TMD, transmembrane domain; IP, inositol phosphate.
References
- 1.Khorana H G. J Biol Chem. 1992;267:1–4. [PubMed] [Google Scholar]
- 2.Savarese T M, Fraser C M. Biochem J. 1992;283:1–19. doi: 10.1042/bj2830001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheer A, Fanelli F, Costa T, De Benedetti P G, Cotecchia S. EMBO J. 1996;15:3566–3578. [PMC free article] [PubMed] [Google Scholar]
- 4.Kjelsberg M A, Cotecchia S, Ostrowski J, Caron M G, Lefkowitz J R. J Biol Chem. 1992;267:1430–1433. [PubMed] [Google Scholar]
- 5.Cotecchia S, Exum S, Caron M G, Lefkowitz R J. Proc Natl Acad Sci USA. 1990;87:2896–2900. doi: 10.1073/pnas.87.8.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly M L. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 7.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 8.Charton M. In: Progress in Physical Organic Chemistry. Taft R W, editor. New York: Wiley; 1990. pp. 163–284. [Google Scholar]
- 9.Hopp T P, Woods K R. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp K A, Nicholls A, Friedman R, Barry H. Biochemistry. 1991;30:9686–9697. doi: 10.1021/bi00104a017. [DOI] [PubMed] [Google Scholar]
- 11.Dawson D M. In: The Biochemical Genetics of Man. Brock D J H, Mayo O, editors. New York: Academic; 1971. pp. 1–38. [Google Scholar]
- 12.Grantham R. Science. 1984;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 13.Krigbaum W R, Komoriya A. Biochim Biophys Acta. 1979;576:204–228. doi: 10.1016/0005-2795(79)90498-7. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson L, Jonsson J, Sjostrom M, Wold S. Quant Struct Act Relat. 1988;7:144–150. [Google Scholar]
- 15.Weber A L, Lacey J C., Jr J Mol Biol. 1978;118:289–304. [Google Scholar]
- 16.Naray-Szabo G. J Mol Graphics. 1989;7:76–98. doi: 10.1016/s0263-7855(89)80003-7. [DOI] [PubMed] [Google Scholar]
- 17.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 18.Eisemberg D, Weiss R M, Terwilliger T C, Wilcox W. Faraday Symp Chem Soc. 1982;17:109–120. [Google Scholar]
- 19.Pliska V, Schmidt M, Fauchere J-L. J Chromatogr. 1981;216:79–92. [Google Scholar]
- 20.Manavalan P, Pannuswamy Nature (London) 1976;275:673–674. doi: 10.1038/275673a0. [DOI] [PubMed] [Google Scholar]
- 21.Chothia C. J Mol Biol. 1976;105:1–14. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 22.Hellberg S, Sjostrom M, Skagerberg B, Wold S. J Med Chem. 1987;30:1126–1135. doi: 10.1021/jm00390a003. [DOI] [PubMed] [Google Scholar]
- 23.Frommel C. J Theor Biol. 1984;111:247–260. doi: 10.1016/s0022-5193(84)80209-x. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira L, Paiva A C M, Sander C, Vriend G. Trends Pharmacol Sci. 1994;15:170–172. doi: 10.1016/0165-6147(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 25.Fanelli F, Menziani M C, De Benedetti P G. Protein Eng. 1995;8:557–564. doi: 10.1093/protein/8.6.557. [DOI] [PubMed] [Google Scholar]
- 26.Cohen G B, Yang T, Robinson P R, Oprian D D. Biochemistry. 1993;32:6111–6115. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- 27.Franke R R, König B, Sakmar T P, Khorana H G, Hofmann K P. Science. 1990;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 28.Arnis S, Fahmy K, Hofmann K P, Sakmar T P. J Biol Chem. 1994;269:23879–23881. [PubMed] [Google Scholar]
- 29.Urry D W, Peng S Q, Parker T M, Gowda D C, Harris R D. Angew Chem Int Ed Engl. 1993;32:1440–1442. [Google Scholar]
- 30.Tributsch H, Pohlmann L. J Theor Biol. 1996;178:17–28. doi: 10.1006/jtbi.1996.0003. [DOI] [PubMed] [Google Scholar]