Abstract
The method of site-directed spin labeling (SDSL) utilizes a stable nitroxide radical to obtain structural and dynamic information on biomolecules. Measuring dipolar interactions between pairs of nitroxides yields internitroxide distances, from which quantitative structural information can be derived. This study evaluates SDSL distance measurements in RNA using a nitroxide probe, designated as R5, which is attached in an efficient and cost-effective manner to backbone phosphorothioate sites that are chemically substituted in arbitrary sequences. It is shown that R5 does not perturb the global structure of the A-form RNA helix. Six sets of internitroxide distances, ranging from 20 to 50 Å, were measured on an RNA duplex with a known X-ray crystal structure. The measured distances strongly correlate (R2 = 0.97) with those predicted using an efficient algorithm for determining the expected internitroxide distances from the parent RNA structure. The results enable future studies of global RNA structures for which high-resolution structural data are absent.
INTRODUCTION
RNA performs many essential cellular functions involving the maintenance, transfer, and processing of genetic information (1). With recent discoveries such as RNA interference and riboswitches, the scope of RNA biological activities keeps expanding. The versatile functions of RNA rely on its ability to adopt diverse three-dimensional structures, and an in-depth understanding of RNA structure and dynamics is key to understanding RNA function. There has been a recent leap in the number of available high-resolution RNA structures, perhaps best exemplified by reports of ribosome structures (2), but studies of RNA structure and dynamics remain challenging. X-ray crystallography is limited by the availability of crystals and the static nature of the information obtained, and NMR spectroscopy is inherently restricted by the size of the nucleic acids. Therefore, it is of interest to explore techniques that are capable of providing structural and dynamic information on arbitrary size systems in solution.
Site-directed spin labeling (SDSL) has matured as a means for studying structure and dynamics of proteins that are difficult to analyze using x-ray crystallography or NMR spectroscopy (3–9). In SDSL, a stable nitroxide radical is covalently attached to a specific site within a macromolecule, and information on the local site is derived via electron paramagnetic resonance (EPR) spectroscopy. Development of SDSL for studying nucleic acids lags behind that of proteins, but the available literature indicates that SDSL is capable of providing structural and dynamic information at the level of individual nucleotides (10).
The most recent advancement in SDSL is to use pulse EPR techniques to measure distances in the nanometer range (11–14). The distance between a pair of electron spins can be measured by monitoring the interspin dipolar interaction. Interspin distances between 5 and 20 Å can be measured using continuous-wave EPR (15–17), and those between 20 and 80 Å are accessible using pulse EPR methods (18–26). The measured interspin distances provide direct structural information and potentially would allow one to map the global structure of the parent molecule. Although the conceptual framework of SDSL distance measurements is similar to that of fluorescence resonance energy transfer (FRET), the nitroxide probes used in SDSL studies are generally smaller than the fluorophores and, therefore, should cause less perturbation to the parent structure.
Recently, nanometer distance measurements in DNA have been reported utilizing a nitroxide that is attached to phosphorothioates that are introduced at specific backbone positions via solid-phase chemical synthesis (26). This nitroxide probe, designated as R5 (Fig. 1), has the advantage that it can be attached, in an efficient and cost-effective manner, to arbitrary DNA sequences. Studies have shown that R5 presents no measurable perturbation to the B-form DNA duplex (26). When one properly accounts for the presence of the two phosphorothioate diastereomers that are introduced during solid-phase synthesis, the measured internitroxide distances have an excellent correlation with predicted values obtained based on the NMR structure of the parent DNA (26). The results indicate that R5 is a universal tag for conducting unrestricted distance measurements that may yield the global structure of arbitrary DNA molecules.
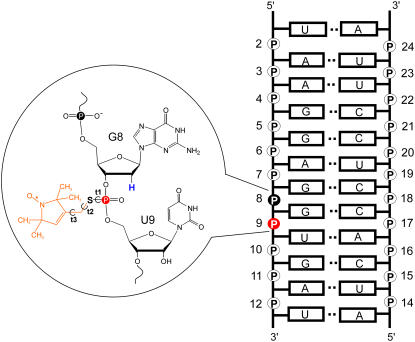
FIGURE 1.
Model SDR RNA. The RNA sequence is shown on the right, with the phosphate positions numbered. By convention each phosphate “belongs” to the ribose that is on the 3′-side of the P atom. The inset shows the detailed chemical structure of an R5 (orange) attached at position 9. Notice the deoxyribose substitution at nucleotide 8, which replaces the 2′-OH group adjacent to the labeled phosphorothioate with a 2′-H (blue).
The R5 probe can also be attached to RNA (27) and is potentially applicable in RNA structural mapping. However, a number of issues need to be examined to confirm that R5 is suitable for measuring distances in RNA. First, in RNA the helices generally adopt the A-form configuration, which has a narrower and deeper major groove as compared to the B-form helix preferred by the DNA. The deep and narrow major groove limits the available space for accommodating the nitroxide and potentially may give rise to more structural perturbation on R5 labeling. Furthermore, in RNA R5 labeling, the 2′-hydroxyl group adjacent to the labeled phosphorothioate has to be removed to prevent strand scission upon covalent attachment of the nitroxide (27) (Fig. 1). This can be most conveniently achieved by replacing the nucleotide 5′ to the labeling site by a deoxyribonucleotide (27) (Fig. 1). The substitution of the 2′-hydroxyl group could lead to perturbation of the native RNA structure, which will add to potential perturbation effects resulting from the presence of the nitroxide.
In this report, R5 was evaluated for nanometer distance measurements in a dodecamer RNA duplex with a known x-ray crystal structure. Six pairs of R5 were examined, with the average internitroxide distances predicted to fall between 20 and 50 Å based on the known structure. The results indicated that the R5 probes do not alter the overall A-form conformation of the RNA duplex. The experimentally measured internitroxide distances, obtained using double electron-electron resonance (DEER) spectroscopy, correlate strongly (R2 = 0.97) to the predicted values based on a search of sterically allowable R5 conformations in the x-ray structure. Overall, the data demonstrate that R5 is useful in long-range distance measurements in RNA, thus establishing a basis for using R5 to map global structures of RNA molecules without known high-resolution structures.
MATERIALS AND METHODS
RNA oligonucleotides and R5 labeling
The sequence of the model RNA duplex, designated as “SDR”, is shown in Fig. 1, with the backbone phosphate positions numbered from 2 to 24. All RNA oligonucleotides were synthesized using solid-phase RNA synthesis (Integrated DNA Technology, Coralville, IA). For RNAs containing a site-specific phosphorothioate modification, the nucleotide 5′ to the phosphorothioate modification was modified to the corresponding 2′-deoxyribonucleotide. When a 2′-deoxyuridine was required, the nucleotide was replaced by a 2′-deoxythymine. The crude synthetic oligonucleotides were reacted with an iodomethyl derivative of a nitroxide following a previously reported procedure (26). The nitroxide-labeled oligonucleotides were purified using anion-exchange HPLC (26). RNA concentrations were determined according to absorbance at 260 nm, using extinction coefficients of 131,300 M−1cm−1 and 112,400 M−1cm−1 for the respective RNA strands and their phosphorothioate and nitroxide-labeled derivatives. All RNAs were stored at −20°C in water.
Characterizations of R5-labeled RNA
Thermal denaturation of RNA duplexes was carried out in a buffer containing 100 mM NaCl and 10 mM sodium phosphate (pH 6.8) following a previously reported procedure (28,29). The standard state free energy of duplex-to-single-strand transition at 37°C (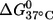 ) was obtained as described (29). Circular dichroism spectra were measured at 20°C between 320 and 205 nm on a JASCO J-715 spectropolarimeter. The duplexes were dissolved in the same buffer as those used for thermal denaturation measurements.
) was obtained as described (29). Circular dichroism spectra were measured at 20°C between 320 and 205 nm on a JASCO J-715 spectropolarimeter. The duplexes were dissolved in the same buffer as those used for thermal denaturation measurements.
Four-pulse DEER spectroscopy
The double-labeled SDR duplexes were formed by mixing the appropriate singly-labeled individual strands in a 1:1 ratio. The RNA mixture was heated at 95°C for 1 min and then cooled at room temperature for 2 min. An appropriate amount of a buffer containing NaCl and Tris (2-amino-2-(hydroxymethyl)-1,3-propanediol, pH 7.5) was added, and the mixture was incubated at room temperature for >5 h and then lyophilized. The SDR duplex was then resuspended in 10 μl, with the final solution containing 100 mM NaCl, 50 mM Tris pH 7.5, 20% (v/v) glycerol, and 90–150 μM double-labeled RNA.
Four-pulse DEER measurements were performed in a Bruker ELEXSYS 580 spectrometer following a previously reported procedure (26). The echo decay data were analyzed using the DeerAnalysis2006 package developed by Jeschke and co-workers (freely available at http://www.mpip-mainz.mpg.de/∼jeschke/distance.html). The distance distribution functions (P(r)) were computed using Tikhonov regularization. The mean distance (〈rDEER〉) and the width of the distance distribution (σDEER) for a selected range were calculated as
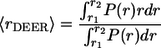 |
(1) |
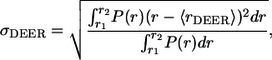 |
(2) |
with r1 representing the lower bound and r2 representing the upper bound. Error in 〈rDEER〉 is estimated to be 5% based on multiple measurements of selective samples.
Computer modeling
Theoretical internitroxide distances were calculated using an in-house conformer-search algorithm, referred to as NASNOX, incorporated within the framework of a larger nucleic acids modeling program, NASDAC (30). The NASNOX algorithm, which has been described in detail elsewhere (31), allows addition of R5 labels at desired phosphorothioate sites of an input nucleic acid structure. Here, the input structure is the x-ray structure of the SDR duplex (Protein Data Bank ID: 1SDR) (32), with deletion of the O2′ atom adjacent to each R5 attachment site. If O2′ deletion is required in an rU nucleotide, the nucleotide was changed to a dT by addition of a C atom to C5 in a standard geometry using WebLab ViewerPro (v. 3.7, Molecular Simulations, San Diego, CA). For all calculations, protons were added by NASNOX in a standard geometry (30,31). The torsion angle S-P-O5′-C5′ (Fig. 1 A) was set at the value in the original duplex (with S substituted for O and a P-S bond of 1.99 Å) and torsion angles t1, t2, and t3 (Fig. 1 A) were varied. The remaining geometry of the label has been described previously (26,31).
The ensemble of allowable R5 conformers was identified by stepwise variation of torsion angles t1, t2, and t3 while fixing the RNA coordinates. The increments (I) were set at 120° for t1 (i.e., torsions of 180°, 60°, and −60°), 120° for t2 (torsions of 180°, 60°, and −60°), and 30° for t3 (12 torsional positions, starting at 0°). Allowable conformations of R5 are defined as those conformers in which no atoms of the nitroxide and RNA are within 75% of the sum of the van der Waals radii of the corresponding atoms. During the search, if a clash between the nitroxide and RNA was registered for a particular set of (t1, t2, t3), a “fine search” of the local conformational space was carried out (31). The “fine search” subroutine evaluates 125 (i.e., 53) sets of modified (t1, t2, t3) generated by independently varying each torsion angle through five finer increments: the original value of torsion angle (T) minus 20% of the corresponding increment (I) (T − 0.2*I); T − 0.1*I; T; T + 0.1*I; and T + 0.2*I. Any modified (t1, t2, t3) set that relieves the clash between the nitroxide and the RNA is identified as a “modified-fit” conformer, and the average of all modified-fit conformers for an original (t1, t2, t3) position is included in the ensemble for calculating the internitroxide distance.
For a given SDR duplex with R5 modeled at two locations, distances were calculated between the nitroxide nitrogen atoms. Corresponding average distances and the widths of the distribution (as characterized by the standard deviation) were calculated based on the ensemble of distances. Calculations were carried out with inclusion of both Rp and Sp diastereomers at each site. In the reported crystal structure (32), the unit cell includes two double-stranded RNA molecules with similar structures. Internitroxide distances were calculated for both structures, and the average distances are reported.
RESULTS
The model system
For evaluating SDSL distance measurements, a dodecamer RNA duplex containing non-self-complementary sequences with Watson-Crick base pairings was chosen as a model system (Fig. 1). The model RNA was designated as SDR, following the Protein Data Bank ID of its x-ray structure, which shows an A-form helix (32). In SDR, ∼40% of the 121 interstrand phosphorus-phosphorus distances are in the range of 17 Å to 40 Å. It is expected that these sites are suitable for conducting DEER measurements.
Double-labeled SDR duplexes were assembled from two singly labeled complementary strands. Anion-exchange HPLC and denaturing gels showed that the efficiency of R5 labeling is close to 100%, and MALDI-TOF mass spectrometry analyses confirmed that each RNA strand has one, and only one, R5 attached (supplemental Fig. S1). RNA duplex formation was confirmed on native gels. In this article, a particular SDR duplex is identified by specifying the positions of the nitroxide labeling sites. For example, in sample (9;17), nucleotides 9 and 17 are modified with phosphorothioates and labeled with R5, and nucleotides 8 and 16 (the corresponding 5′ nucleotides) are replaced by deoxyribonucleotides. Phosphorothioate substitution via solid-phase chemical synthesis yields two diastereomers (Rp and Sp) with a ∼50/50 ratio. The diastereomers were not separated in the studies reported here.
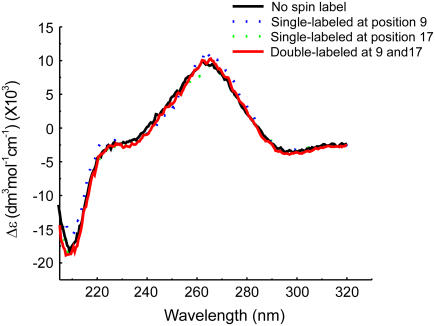
Circular dichroism (CD) spectroscopy was used to access structural perturbations in the nitroxide-labeled RNA duplexes. The CD spectra of the unlabeled and the double-labeled SDR duplexes are very similar (Fig. 2). The CD spectra showed a positive band centered at ∼268 nm and a negative band around ∼210 nm, which are characteristic of A-form RNA (33). The results indicate that the attachment of the nitroxide and the substitution of a deoxyribonucleotide 5′ to the labeling site did not globally alter the RNA duplex structure.
FIGURE 2.
Representative set of CD spectra of R5-labeled SDR RNA duplexes. The “no spin label” spectrum is obtained from an all-ribose SDR duplex without any phosphorothioate or deoxyribose modification. For spectra of labeled SDR duplexes, the positions of R5 are indicated in parentheses.
The nitroxide-labeled duplexes were further characterized by thermal melting measurements (Table 1). For the (12;24) duplex, which has the nitroxides attached near the termini, no change was observed in 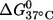 between unlabeled and double-labeled duplexes, indicating no perturbation caused by nitroxide labeling. For the other duplexes,
between unlabeled and double-labeled duplexes, indicating no perturbation caused by nitroxide labeling. For the other duplexes, 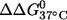 ranged from 1.6 to 3.3 kcal/mol. The largest
ranged from 1.6 to 3.3 kcal/mol. The largest 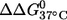 decrease (3.3 kcal/mol, Table 1) corresponds to ∼10°C decrease in the melting temperature. A similar decrease of melting temperature has been reported (17), where SDSL distance measurements were carried out on an RNA duplex using a pair of spin labels attached to the 2′ position of the ribose.
decrease (3.3 kcal/mol, Table 1) corresponds to ∼10°C decrease in the melting temperature. A similar decrease of melting temperature has been reported (17), where SDSL distance measurements were carried out on an RNA duplex using a pair of spin labels attached to the 2′ position of the ribose.
TABLE 1.
Thermodynamic parameters for R5-labeled SDR duplexes
| Nitroxide positions |
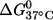 (kcal/mol) (kcal/mol) |
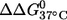 (kcal/mol) (kcal/mol) |
|---|---|---|
| Unlabeled* | 13.5 | |
| (12;24) | 13.5 | 0.0 |
| (11;23) | 11.9 | 1.6 |
| (12;19) | 11.2 | 2.3 |
| (9;23) | 11.2 | 2.3 |
| (9;24) | 10.9 | 2.6 |
| (9;17) | 10.2 | 3.3 |
| (9;–)† | 12.3 | 1.2 |
| (–;17)† | 13.0 | 0.5 |
All-ribose SDR duplex without any phosphorothioate or deoxyribose modification.
“–” indicates no R5 label at the corresponding strand.
It is noted that the (9;17) duplex, which shows the largest 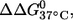 has very similar CD spectra between unlabeled and double-labeled duplexes (Fig. 2), suggesting that the difference in
has very similar CD spectra between unlabeled and double-labeled duplexes (Fig. 2), suggesting that the difference in 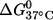 might result from perturbations localized to the labeling sites. Further melting studies showed that the singly labeled (9;–) and (–;17) duplex each have a small
might result from perturbations localized to the labeling sites. Further melting studies showed that the singly labeled (9;–) and (–;17) duplex each have a small 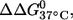 and the sum is smaller than the measured
and the sum is smaller than the measured 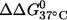 for the double-labeled duplex (9;17) (Table 1). This suggests a cooperative effect between the two nitroxides, which might occur because positions 9 and 17 are located directly across from each other, and the nitroxides might be perturbing the same basepair.
for the double-labeled duplex (9;17) (Table 1). This suggests a cooperative effect between the two nitroxides, which might occur because positions 9 and 17 are located directly across from each other, and the nitroxides might be perturbing the same basepair.
Overall, the data indicate that double nitroxide labeling does not alter the global A-form helix structure, but may locally perturb the parent structure. This is similar to the situation in spin-labeled proteins, where introduction of a spin label at some sites destabilizes the protein but does not significantly alter the structure of the protein (34).
DEER measurements in RNA using R5
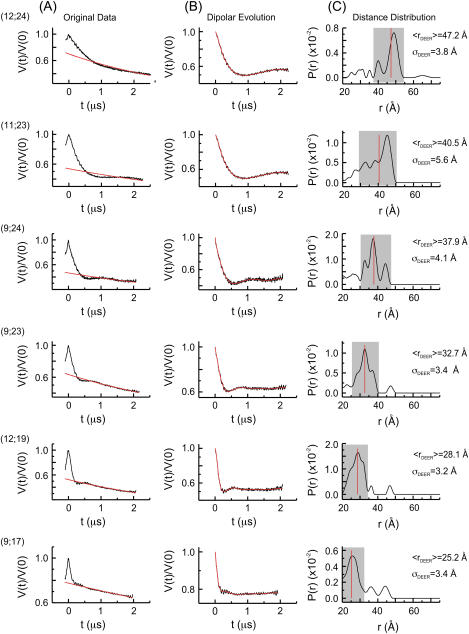
Application of the four-pulse DEER has been described for a pair of R5′s attached to DNA (26). Following similar protocols (see Materials and Methods), SDR duplexes with a pair of R5′s attached to specific sites were assembled. Dipolar evolution functions were measured, and the corresponding distance distribution probabilities (P(r)) were computed via Tikhonov regularization. For the six sets of measured internitroxide distances, the resulting average distances (〈rDEER〉) range from 25.2 Å to 47.2 Å (Table 2 and Fig. 3). The longest distance measured, 〈rDEER(12;24)〉 = 47.2 Å (Fig. 3), is between a pair of R5 at sites 12 and 24, which are located at the 3′ termini of the individual SDR strands. This is ∼10 Å longer than the longest reported DEER measured distance in an R5-labeled DNA duplex (26) and reflects the nature of the A-form RNA duplex.
TABLE 2.
Correlation between measured and predicted average distances
| Nitroxide positions | 〈rDEER〉 (Å)* | 〈rmodel〉 (Å)† |
|---|---|---|
| (9;17) | 25.2 | 24.5 |
| (12;19) | 28.1 | 31.1 |
| (9;23) | 32.7 | 34.7 |
| (9;24) | 37.9 | 38.8 |
| (11;23) | 40.5 | 42.4 |
| (12;24) | 47.2 | 46.6 |
| RMSD (Å) | 1.7 | |
| R2 | 0.97 | |
Measured from the major population marked by the shaded boxes in Fig. 3 C.
Computed using NASNOX with the “fine search” procedure described in Materials and Methods.
FIGURE 3.
DEER data. The positions of R5 are shown in parentheses. (A) Original echo decay data. The black traces are the measured echo amplitude that has been normalized to the amplitude at t = 0. The red traces are the background echo decay computed using a homogeneous three-dimensional spin distribution. (B) Dipolar evolution functions. The black traces represent the differences between the measured echo decay and the background decay shown in A. The red traces are the simulated echo decay computed according to the corresponding distance distributions shown in C. (C) The distance distributions (P(r)), which were computed with a regularization parameter of 10. Varying the regularization parameter from 1 to 100 did not significantly change the computed P(r). The major band in each P(r), defined as those distances with P(r) ≥ (0.25 × Pmax), is marked by a shaded box. The red lines mark the average distances calculated for each major band.
The widths of the distance distribution, characterized by the standard deviations (σDEER), range from 3.2 Å to 5.6 Å (Table 3). These are larger than those observed in the DNA duplex (1.9 Å to 5.4 Å (26)) and reflect that P(r) for the SDR duplex is broad and contains multiple peaks. Our current method of analysis collapses P(r) to a single average distance 〈rDEER〉, which is calculated from one major band distributed continuously along the distance axis (indicated by the shaded box, Fig. 3). The analysis excludes minor peaks, which in this work were defined as those with P(r) < 25% of the maximum P (Fig. 3). In every computed P(r), all minor peaks are located outside of the major band and do not disrupt the continuity of the major band. They may represent minor RNA populations or may arise as a result of artifacts in data fitting (35). Excluding these minor peaks does not significantly alter the 〈rDEER〉 values (supplemental Table S1).
TABLE 3.
Measured and predicted widths of distance distributions
| Nitroxide positions | 〈σDEER〉 (Å)* | 〈σmodel〉 (Å)† |
|---|---|---|
| (9;17) | 3.4 | 3.0 |
| (12;19) | 3.2 | 2.6 |
| (9;23) | 3.4 | 2.7 |
| (9;24) | 4.1 | 3.1 |
| (11;23) | 5.6 | 3.1 |
| (12;24) | 3.8 | 2.3 |
Measured from the major population marked by the shaded boxes in Fig. 3 C.
Computed using NASNOX with the “fine search” procedure described in Materials and Methods.
Correlations between measured and expected distances
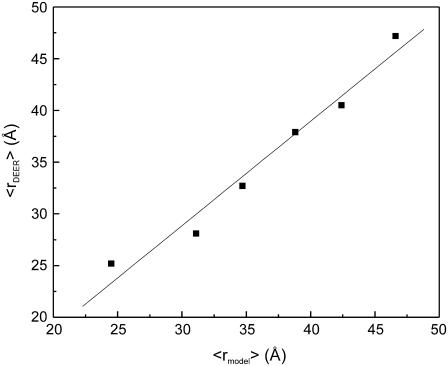
The measured 〈rDEER〉 was compared to expected mean distances (〈rmodel〉, Table 2), which were computed using the NASNOX algorithm as described in Materials and Methods (Fig. 4). The result showed that 〈rDEER〉 correlates with 〈rmodel〉 with an overall root mean-square deviation (RMSD) of 1.7 Å (Table 2). When 〈rDEER〉 was plotted as a function of 〈rmodel〉, the data fit to 〈rDEER〉 = 1.0 × 〈rmodel〉 − 1.4 Å, with a correlation coefficient (R2) of 0.97 (Fig. 5). The fit has a slope of unity and an offset that is small compared to the distances measured. Such a good correlation between 〈rDEER〉 and 〈rmodel〉 suggests that R5 can be used to reliably measure distances in RNA.
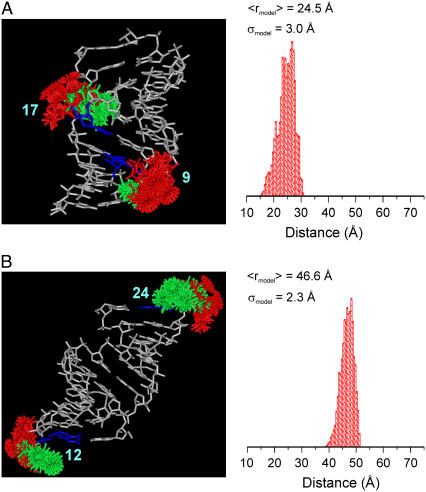
FIGURE 4.
Ensembles of allowable R5 conformations modeled at sites (9;17) (A) and sites (12;24) (B) of the SDR RNA. The RNA is shown in white, with the labeled nucleotides shown in blue. Conformers attached to the Sp diastereomer are shown in red, and those attached to the Rp diastereomer are in green. On the right is a histogram of the predicted distance distribution, with the average distance (〈rmodel〉) and width of the distribution (σmodel) shown.
FIGURE 5.
Correlation between the measured (〈rDEER〉) and predicted (〈rmodel〉) average distances. The solid line represents a linear fit of 〈rDEER〉 = 1.0 × 〈rmodel〉 − 1.4 Å.
The measured standard deviations (σDEER) are all larger than the predicted values (σmodel) (Table 3). Deviation of σDEER from σmodel has been observed in the DNA study (26) and may be related to the fact that the coordinate of the RNA is fixed in the search algorithm. A number of predicted σmodel in the RNA are larger than those in the DNA, which is consistent with the broader measured distance distribution (vide supra). For example, σmodel(9;17) (direct cross-strand distance) is 3.0 Å in RNA (Table 3), but its counterpart in DNA is 1.5 Å (σ(7;19) from Cai et al. (26)). This is consistent with the measured σDEER(9;17) in RNA (3.4 Å) being larger than its counterpart in DNA (1.9 Å) (26). Therefore, both the DEER measurement and the NASNOX algorithm report the difference between the A-form and B-form duplex.
The “fine search” option in the NASNOX algorithm (see Materials and Methods) lessens the problem of identifying R5 conformers in the deep and narrow major groove of the A-form RNA duplex. This provides better coverage of the allowable R5 conformational space. As expected, more R5 conformers are identified when the “fine search” function is invoked, and the 〈rDEER〉/〈rmodel〉 correlation is slightly better (supplemental Table S2).
DISCUSSION
Work presented here demonstrates that the R5 nitroxide probe can be used to measure nanometer distances in RNA molecules. These results complement a previous report on measuring distances in DNA utilizing R5 (26). Together, these studies establish the applicability of R5 for mapping structures of nucleic acids.
The R5 probe has a number of features that are advantageous for structural mapping in nucleic acids. It utilizes a universal functional substitution in nucleic acids and therefore can be attached to arbitrary sequences. The chemistry for R5 labeling, from the introduction of the phosphorothioate to the covalent addition of the nitroxide moiety, is highly efficient. The cost of R5 labeling is ∼10-fold less than those of other methods (17,20,28,36–40). With these features, it is possible to “scan” the R5 probe through the sequence of a DNA or RNA and obtain multiple distance constraints. This is key in mapping global structures.
R5 is a nonnative functionality of nucleic acids and is subject to the limitations of an extrinsic probe, mainly potential perturbations to the native structure. There are two most likely sources of perturbation on R5 labeling in RNA: 1), the deep and narrow major grove of the A-form helix, which gives limited space to accommodate the R5 probe; and 2), the required removal of the 2′-OH group adjacent to the labeling site. Data presented above reveal an excellent correlation between the measured and predicted average distances as well as standard A-form helix CD spectra in the double-labeled RNAs. These indicate that R5 does not severely perturb the global structure of an RNA helix. On the other hand, reductions of 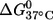 were observed in melting studies of certain double-labeled RNAs, suggesting that the R5 label may, to a certain degree, perturb the local environment at certain labeling sites. It is noted that perturbations caused by an extrinsic probe, including R5, are system specific and need to be examined case by case. Data reported here suggest one can selectively attach R5 within duplex regions in mapping RNA global structures, thus minimizing potential disruptions of the native structure.
were observed in melting studies of certain double-labeled RNAs, suggesting that the R5 label may, to a certain degree, perturb the local environment at certain labeling sites. It is noted that perturbations caused by an extrinsic probe, including R5, are system specific and need to be examined case by case. Data reported here suggest one can selectively attach R5 within duplex regions in mapping RNA global structures, thus minimizing potential disruptions of the native structure.
Correlating the measured internitroxide distances to those of the parent molecule is another challenge. In this study, the conformer search method (NASNOX), which was originally developed in DNA studies (26,31), was shown to be applicable to RNA. With the improved fine-search option, NASNOX is able to accurately predict the internitroxide distances based on the available x-ray structure. The key advantage of NASNOX is its speed: it can predict an internitroxide distance within a couple of minutes on a PC, as compared to weeks of computation time required for MD simulations (31). In addition, NASNOX can simultaneously account for the presence of the two phosphorothioate diastereomers at both labeling sites. This is in contrast to the MD simulations, where four separate simulations are required to account for the diastereomers and yield the predicted distances (31). The limitation in NASNOX is that the coordinates of the nucleic acids are fixed, and therefore, the algorithm is most useful in cases where the RNA structures are not altered by the nitroxide probe.
In conclusion, data presented here establish two basic components for mapping global RNA structures utilizing R5: 1), experimental measurements of nanometer distances; and 2), an efficient algorithm for correlating the measured internitroxide distances to the parent RNA structure. This approach will enable future studies of RNA without prior high-resolution structures.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Dr. Wayne L. Hubbell and Dr. Glenna Sowa for thoughtful comments on the manuscript. We thank Dr. Hubbell for providing access to a Bruker E-580 spectrometer.
Research reported was supported by a National Science Foundation Career award to P.Z.Q. (MCB 054652). K.H. was supported by a grant from the Hungarian National Research Fund (OTKA T48334).
Editor: David D. Thomas.
References
- 1.Gesteland, R. F., J. F. Atkins, and T. R. Cech, editors. 2006. RNA World, 3rd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- 2.Moore, P. B., and T. A. Steitz. 2005. The ribosome revealed. Trends Biochem. Sci. 30:281–283. [DOI] [PubMed] [Google Scholar]
- 3.Hubbell, W. L., A. Gross, R. Langen, and M. A. Leitzow. 1998. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 8:649–656. [DOI] [PubMed] [Google Scholar]
- 4.Feix, J. B., and C. S. Klug. 1998. Site-directed spin labeling of membrane proteins and peptide-membrane interactions. In Biological Magnetic Resonance. L. J. Berliner, editor. Plenum Press, New York. 251–281.
- 5.Hubbell, W. L., D. S. Cafiso, and C. Altenbach. 2000. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 7:735–739. [DOI] [PubMed] [Google Scholar]
- 6.Fajer, P. G. 2000. Electron spin resonance spectroscopy labeling in proteins and peptides analysis. In Encyclopedia of Analytical Chemistry. R. Meyers, editor. John Wiley & Sons, Chichester. 5725–5761.
- 7.Columbus, L., and W. L. Hubbell. 2002. A new spin on protein dynamics. Trends Biochem. Sci. 27:288–295. [DOI] [PubMed] [Google Scholar]
- 8.Hubbell, W. L., C. Altenbach, C. M. Hubbell, and H. G. Khorana. 2003. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv. Protein Chem. 63:243–290. [DOI] [PubMed] [Google Scholar]
- 9.Fanucci, G. E., and D. S. Cafiso. 2006. Recent Advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 16:644–653. [DOI] [PubMed] [Google Scholar]
- 10.Qin, P. Z., and T. Dieckmann. 2004. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Struct. Biol. 14:350–359. [DOI] [PubMed] [Google Scholar]
- 11.Borbat, P. P., A. J. Costa-Filho, K. A. Earle, J. K. Moscicki, and J. H. Freed. 2001. Electron spin resonance in studies of membranes and proteins. Science. 291:266–269. [DOI] [PubMed] [Google Scholar]
- 12.Lakshmi, K. V., and G. W. Brudvig. 2001. Pulsed electron paramagnetic resonance methods for macromolecular structure determination. Curr. Opin. Struct. Biol. 11:523–531. [DOI] [PubMed] [Google Scholar]
- 13.Jeschke, G. 2005. EPR techniques for studying radical enzymes. Biochim. Biophys. Acta. 1707:91–102. [DOI] [PubMed] [Google Scholar]
- 14.Jeschke, G. 2002. Distance measurements in the nanometer range by pulse EPR. Chem. Phys. Chem. 3:927–932. [DOI] [PubMed] [Google Scholar]
- 15.Rabenstein, M. D., and Y. K. Shin. 1995. Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. USA. 92:8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altenbach, C., K. J. Oh, R. J. Trabanino, K. Hideg, and W. L. Hubbell. 2001. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry. 40:15471–15482. [DOI] [PubMed] [Google Scholar]
- 17.Kim, N., A. Murali, and V. J. DeRose. 2004. A distance ruler for RNA using EPR and site-directed spin labeling. Chem. Biol. 11:939–948. [DOI] [PubMed] [Google Scholar]
- 18.Persson, M., J. R. Harbridge, P. Hammarstrom, R. Mitri, L. G. Martensson, U. Carlsson, G. R. Eaton, and S. S. Eaton. 2001. Comparison of electron paramagnetic resonance methods to determine distances between spin labels on human carbonic anhydrase II. Biophys. J. 80:2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borbat, P. P., H. S. McHaourab, and J. H. Freed. 2002. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J. Am. Chem. Soc. 124:5304–5314. [DOI] [PubMed] [Google Scholar]
- 20.Schiemann, O., A. Weber, T. E. Edwards, T. F. Prisner, and S. T. Sigurdsson. 2003. Nanometer distance measurements on RNA using PELDOR. J. Am. Chem. Soc. 125:3334–3335. [DOI] [PubMed] [Google Scholar]
- 21.Schiemann, O., N. Piton, Y. Mu, G. Stock, J. W. Engels, and T. F. Prisner. 2004. A PELDOR based nanometer distance ruler for oligonucleotides. J. Am. Chem. Soc. 126:5722–5729. [DOI] [PubMed] [Google Scholar]
- 22.Borbat, P. P., J. H. Davis, S. E. Butcher, and J. H. Freed. 2004. Measurement of large distances in biomolecules using double-quantum filtered refocused electron spin-echoes. J. Am. Chem. Soc. 126:7746–7747. [DOI] [PubMed] [Google Scholar]
- 23.Piton, N., O. Schiemann, Y. Mu, G. Stock, T. Prisner, and J. W. Engels. 2005. Synthesis of spin-labeled RNAs for long range distance measurements by peldor. Nucleosides Nucleotides Nucleic Acids. 24:771–775. [DOI] [PubMed] [Google Scholar]
- 24.Sale, K., L. Song, Y. S. Liu, E. Perozo, and P. Fajer. 2005. Explicit treatment of spin labels in modeling of distance constraints from dipolar EPR and DEER. J. Am. Chem. Soc. 127:9334–9335. [DOI] [PubMed] [Google Scholar]
- 25.Banham, J. E., C. R. Timmel, R. J. Abbott, S. M. Lea, and G. Jeschke. 2006. The characterization of weak protein-protein interactions: evidence from DEER for the trimerization of a von Willebrand Factor A domain in solution. Angew. Chem. Int. Ed. Engl. 45:1058–1061. [DOI] [PubMed] [Google Scholar]
- 26.Cai, Q., A. K. Kusnetzow, W. L. Hubbell, I. S. Haworth, G. P. Gacho, N. Van Eps, K. Hideg, E. J. Chambers, and P. Z. Qin. 2006. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 34:4722–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin, P. Z., S. E. Butcher, J. Feigon, and W. L. Hubbell. 2001. Quantitative analysis of the GAAA tetraloop/receptor interaction in solution: A site-directed spin labeling study. Biochemistry. 40:6929–6936. [DOI] [PubMed] [Google Scholar]
- 28.Qin, P. Z., K. Hideg, J. Feigon, and W. L. Hubbell. 2003. Monitoring RNA base structure and dynamics using site-directed spin labeling. Biochemistry. 42:6772–6783. [DOI] [PubMed] [Google Scholar]
- 29.Qin, P. Z., I. Jennifer, and A. Oki. 2006. A model system for investigating lineshape/structure correlations in RNA site-directed spin labeling. Biochem. Biophys. Res. Commun. 343:117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chambers, E. J., E. A. Price, M. Z. Bayramyan, and I. S. Haworth. 2003. Computation of DNA backbone conformations. J. Biomol. Struct. Dyn. 306:177–185. [DOI] [PubMed] [Google Scholar]
- 31.Price, E. A., B. T. Sutch, Q. Cai, P. Z. Qin, and I. S. Haworth. 2007. Computation of nitroxide-nitroxide distances for spin-labeled DNA duplexes. Biopolymers. 87:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin, H., M. Zhang, R. Bald, J. Furste, V. A. Erdmann, and U. Heinemann. 1995. Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol. 249:595–603. [DOI] [PubMed] [Google Scholar]
- 33.Tinoco, I. J., K. Sauer, J. C. Wang, and J. D. Puglisi. 2002. In Physical Chemistry: Principles and Applications in Biological Sciences. Prentice Hall, Upper Saddle River, NJ. 574.
- 34.Mchaourab, H. S., M. A. Lietzow, K. Hideg, and W. L. Hubbell. 1996. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 35:7692–7704. [DOI] [PubMed] [Google Scholar]
- 35.Jeschke, G., G. Panek, A. Godt, A. Bender, and H. Paulsen. 2004. Data analysis procedures for pulse ELDOR measurements of broad distance distributions. Appl. Magn. Reson. 26:223–244. [Google Scholar]
- 36.Edwards, T. E., T. M. Okonogi, B. H. Robinson, and S. T. Sigurdsson. 2001. Site-specific incorporation of nitroxide spin-labels into internal sites of the TAR RNA. Structure-dependent dynamics of RNA by EPR spectroscopy. J. Am. Chem. Soc. 123:1527–1528. [DOI] [PubMed] [Google Scholar]
- 37.Kao, S. C., and A. M. Bobst. 1985. Local base dynamics and local structural features in RNA and DNA duplexes. Biochemistry. 24:5465–5469. [DOI] [PubMed] [Google Scholar]
- 38.Spaltenstein, A., B. H. Robinson, and P. B. Hopkins. 1989. Sequence- and structure-dependent DNA base dynamics: Synthesis, structure, and dynamics of site and sequence specifically spin-labeled DNA. Biochemistry. 28:9484–9495. [DOI] [PubMed] [Google Scholar]
- 39.Gannett, P. M., E. Darian, J. Powell, E. M. Johnson, C. Mundoma, N. L. Greenbaum, C. M. Ramsey, N. S. Dalal, and D. E. Budil. 2002. Probing triplex formation by EPR spectroscopy using a newly synthesized spin label for oligonucleotides. Nucleic Acids Res. 30:5328–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin, P. Z., J. Feigon, and W. L. Hubbell. 2005. Site-directed spin labeling studies reveal solution conformational changes in a GAAA tetraloop receptor upon Mg2+-dependent docking of a GAAA tetraloop. J. Mol. Biol. 351:1–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.