Abstract
The importance of intramolecular disulfides in a noncovalent dimeric protein interleukin-8 (IL-8) has been studied by replacing cysteines in each of the two disulfide pairs with α-aminobutyric acid (CH2-SH → CH2-CH3). Both disulfide mutants are less stable and exist as molten globules in the monomeric state. Interestingly, both mutants dimerize, though with slightly lower affinities compared to the native protein. NMR studies suggest a molten globule-like structure also in the dimeric state. Structures, sequence analysis, and mutagenesis studies have shown that the conserved hydrophobic residues are packed against each other in the protein core and that H bonding and van der Waals interactions stabilize the dimer interface. Deleting either disulfide in IL-8 results in substantial loss in receptor activity, indicating that both disulfides are critical for function in the folded protein. These data together suggest that the packing interactions of the hydrophobic core determine IL-8 monomer fold, that disulfides play only a marginal role in dimer formation, and that the stability imparted by the disulfides is intimately coupled to fold and function.
INTRODUCTION
Since the classical studies of Anfinsen on ribonuclease now more than 40 years ago (1), the role of disulfides has been extensively studied in a variety of monomeric proteins from small peptides to large enzymes. These studies have shown that disulfides are essential and intimately linked to stability, folding, tertiary structure, and function (2–5). However, very little is known regarding how intramolecular disulfides influence quaternary structure formation, and what their role is in the relationship between structure and stability at the monomer and dimer levels.
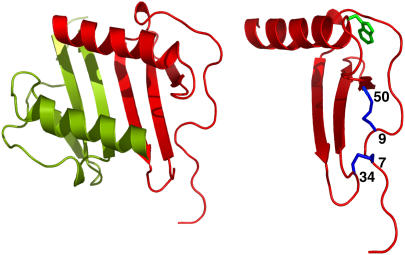
Two characteristic features of chemokine family of proteins are the conserved disulfides and their ability to reversibly associate as dimers. The chemokine interleukin-8 (IL-8) dimerizes with micromolar affinity (6). Structures of native dimer and of a trapped monomer are known and reveal that the monomer structure is essentially similar to that of the monomeric unit in the dimer structure (7,8). Structure-function studies show that both monomeric and dimeric forms play distinct functional roles and that the disulfides are essential for activity (9–12). The structure reveals that the two disulfides (Cys7-Cys34 and Cys9-Cys50) are buried and are located away from the dimer interface (Fig. 1). The 7–34 disulfide is located at the edge of the protein and is not involved in long-range interactions but is nevertheless largely solvent inaccessible. On the other hand, the 9–50 disulfide is in the more structured region of the protein, is involved in long-range interactions, and is also solvent inaccessible.
FIGURE 1.
Schematic of IL-8 dimer and monomer. The monomer structure is shown at a different orientation to highlight the tryptophan and the disulfides.
We report here the consequence of deleting the individual disulfides on IL-8 structure and stability. Compared to the native protein, both disulfide mutants dimerize with slightly lower affinity but are less stable and exist as molten globules in both the monomeric and dimeric states. On the basis of these data, we propose that the packing interactions of the hydrophobic core determine the IL-8 fold and are sufficient for dimer formation and that disulfides stabilize the fold that is essential for function.
MATERIALS AND METHODS
Design and synthesis of IL-8 variants
The recombinant IL-8 (1–66) monomer was produced as described previously (13). This IL-8 variant was used instead of full-length (1–72) IL-8 because it has comparable biological activity and is monomeric at millimolar concentrations. Both IL-8 7–34 and 9–50 disulfide α-aminobutyric acid (Aba) (CH2-SH → CH2-CH3) mutants were produced using solid-phase peptide synthesis and purified by reversed-phase HPLC as described previously (9).
Sedimentation equilibrium
Sedimentation equilibrium experiments were performed using a Beckman-Coulter model XLA analytical ultracentrifuge (Beckman-Coulter, Fullerton, CA) with An-60 titanium rotors using absorbance optics at 280 nm. Experiments were carried using two different loading concentrations of 0.3 and 1.0 mg/ml at rotor speeds 25,000, 30,000, and 35,000 rpm. Sample and blank solutions were made using the same buffer and IL-8 stock solutions by adding a fixed volume of either IL-8 stock solution (sample) or buffer solution (blank). Sedimentation data were fitted to the monomer-dimer equilibrium using the Microcal Origin 4.1 software provided by the manufacturer.
NMR spectroscopy
The NMR data were acquired on a Varian Unity 750 MHz spectrometer (Varian, Palo Alto, CA) equipped with field gradient accessories at 30°C. The mixing time in the nuclear Overhauser effect spectroscopy (NOESY) was 150 ms. The protein concentration was ∼0.5 mM in 20 mM sodium acetate buffer containing 1 mM sodium azide, 1 mM sodium 2,2-dimethyl-2-silapentane sulfonate (DSS), pH 5.2 in 90% H2O/10% 2H2O (v/v). All spectra were referenced to DSS.
Unfolding measurements
The stability of the disulfide deletion mutants was determined by calculating the free energy of unfolding (ΔGF-U) using guanidine hydrochloride (GdnHCl) as a denaturant and fluorescence spectroscopy at 23°C. IL-8 concentration was constant at ∼5 μM, and GdnHCl was increased from 0 to 7 M. The protein concentration was determined using the BCA protein assay (BioRad, Hercules, CA). Sample and blank solutions were made using the same buffer and IL-8 stock solutions by adding a fixed volume of either IL-8 concentrated stock solution (sample) or buffer (blank). The IL-8 spectra were measured on a Spex Fluoromax fluorimeter (Horiba Jobin Yvon, Edison, NJ), with excitation at 295 nm and emission measured from 300 to 400 nm. Blank spectra were measured and subtracted from sample measurements. We also collected the same set of data at 15°C and observed a similar trend in all of the values (not shown). ΔGF-U was obtained by fitting curves using the linear extrapolation method (14).
8-Anilino-1-naphthalene-sulfonic acid as probe for molten globule state
For the 8-anilino-1-naphthalene-sulfonic acid (ANS) fluorescence measurements, IL-8 variants containing different concentrations of GdnHCl were prepared as before, and ANS was added to each sample to a final concentration of 200 μM. ANS fluorescence was measured with excitation at 380 nm, and emission from 400 to 600 nm at 23°C. Control spectra of ANS fluorescence in the presence of GdnHCl were also recorded and subtracted from the protein spectra.
RESULTS AND DISCUSSION
Design of disulfide mutants
The role of the disulfide bond is best studied with cysteines in the reduced state, which in reality is not feasible because disulfide bond formation is highly thermodynamically favored. Therefore, cysteines are replaced by amino acids such as serine and alanine, and if the protein is chemically synthesized, they can also be replaced by isostructural nonnatural amino acids such as Aba. For our studies, we generated two disulfide mutants, one containing Aba substitutions for the Cys7-Cys34 pair and the other for the Cys9-Cys50 pair.
Dimerization of disulfide mutants
The monomer-dimer equilibrium constant (Kd) of the disulfide deletion mutants was measured by the sedimentation equilibrium method (Fig. 2). Native IL-8 dimerizes with micromolar affinity (6), and the dimerization constants for both mutants in phosphate buffer at pH 7 are ∼5-fold lower compared to the wild type (WT) (Table 1). Solution conditions such as pH and ionic strength could influence IL-8 dimerization, so we also measured the Kd in acetate buffer at pH 5.2. The dimerization constants in acetate buffer were also observed to be only slightly lower, indicating that dimerization of the disulfide mutants is not significantly affected by solution conditions (Table 1).
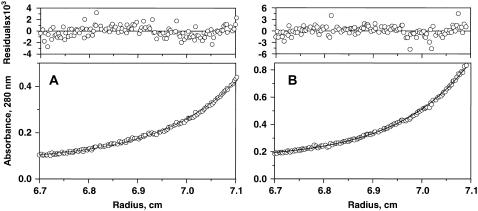
FIGURE 2.
Sedimentation equilibrium data of the (A) 7–34 and (B) 9–50 disulfide mutants in 50 mM sodium phosphate, 50 mM NaCl buffer, pH 7. The concentration in absorbance units is plotted versus r. Residuals of the corresponding least-squares fit are random, indicating the goodness of the fit.
TABLE 1.
Sedimentation equilibrium studies of IL-8 7–34 and 9–50 disulfide mutants
| Protein | Buffer | Kd (μM)* |
|---|---|---|
| WT IL-8 | 50 mM phosphate, pH 7 | 12 ± 5 |
| 50 mM acetate, pH 5.2 | 6 ± 3 | |
| 7–34 disulfide mutant | 50 mM phosphate, pH 7 | 53 ± 12 |
| 50 mM acetate, pH 5.2 | 19 ± 9 | |
| 9–50 disulfide mutant | 50 mM phosphate, pH 7 | 58 ± 9 |
| 50 mM acetate, pH 5.2 | 24 ± 4 |
The Kd reported for data collected at rotor speed of 30,000 rpm. Similar values were obtained at 25,000 and 35,000 rpm, and global fit of the data at all three speeds did not significantly improve the fit. The phosphate buffer also contains 50 mM NaCl
Stability of mutants in the monomeric state
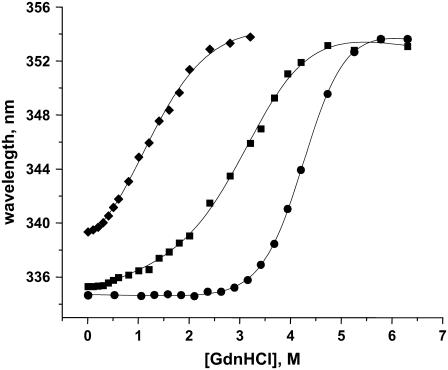
The stabilities of the disulfide mutants were characterized by measuring the free energies of unfolding (ΔGF-U) using GdnHCl denaturation measurements. Both deletion mutants are predominantly monomers under conditions used for fluorescence experiments. However, native IL-8 dimerizes with a Kd ≈ 10 μM, so a fraction of native IL-8 could exist as dimers under conditions used for fluorescence experiments. Therefore, to provide an appropriate monomer control, we used an IL-8 (1–66) deletion mutant that is missing the last six residues. The structure and activity of this IL-8 (1–66) deletion mutant are essentially similar to those of the trapped full-length monomer (13). We made use of the single tryptophan fluorescence (λmax 334 nm) as a probe, which is red-shifted by as much as ∼20 nm on unfolding. The IL-8 structure reveals that the tryptophan is located at the beginning of the α-helix, is buried (accessible surface area 10%), and is in van der Waals contact with a number of residues in the protein core (Fig. 1). Therefore, IL-8 tryptophan fluoresence should serve as a sensitive probe for the structural differences between the WT and the disulfide mutants and for structural changes on unfolding. The λmax of 335 nm for the 7–34 disulfide mutant is similar to that of the IL-8 (1–66) monomer, whereas the 9–50 disulfide mutant is significantly red-shifted, suggesting that the latter mutant should be less structured. On unfolding, all of the proteins show a λmax of 354 nm, which is characteristic of a solvent-exposed tryptophan.
IL-8 (1–66) monomer and both disulfide mutants exhibit a cooperative two-state unfolding transition (Fig. 3). The curves were fit using the linear extrapolation method to derive the midpoint of transition (C½) and ΔG; a summary of these parameters is given in Table 2. Both disulfide mutants are less stable, and the observed ΔGU-F indicates that of the two disulfides, the 9–50 disulfide bond contributes more to the overall global stability of the protein. These observations are consistent with the IL-8 structure, which reveals that the 7–34 disulfide is located at the edge of the protein and is not involved in long-range interactions, whereas the 9–50 disulfide is buried in the more structured region of the protein and is involved in long-range interactions.
FIGURE 3.
Unfolding isotherms of WT (•), 7–34 (▪), and 9–50 (♦) disulfide mutants. The plots show the λmax of tryptophan fluorescence against GdnHCl concentration, and all spectra were collected in 50 mM sodium phosphate, 50 mM NaCl buffer, and pH 7.
TABLE 2.
Equilibrium unfolding studies of IL-8 7–34 and 9–50 disulfide mutants
| Protein | Buffer | ΔG (kcal/mol) | C½ (M) |
|---|---|---|---|
| Native IL-8 | 50 mM phosphate, pH 7.0 | −6.0 ± 0.1 | 4.3 ± 0.1 |
| 50 mM acetate, pH 5.2 | −7.0 ± 0.1 | 3.9 ± 0.1 | |
| 7–34 disulfide mutant | 50 mM phosphate, pH 7.0 | −3.2 ± 0.5 | 3.4 ± 0.1 |
| 50 mM acetate, pH 5.2 | −2.7 ± 0.7 | 3.0 ± 0.2 | |
| 9–50 disulfide mutant | 50 mM phosphate, pH 7.0 | −2.2 ± 0.5 | 1.5 ± 0.2 |
| 50 mM acetate, pH 5.2 | −2.1 ± 0.2 | 1.5 ± 0.1 |
We also measured the unfolding measurements in acetate buffer, pH 5.2, and observed that the stabilities of the mutants are essentially similar in both buffers (Table 2). The differences in steepness of the unfolding transition between the mutants and WT correlate to the m-values of each transition, and these differences empirically indicate a change in accessible surface area between the folded and unfolded states of the mutants in comparison to WT (14). The differences in the shape and baselines of the curves indicate that the two mutants are less stable in the folded state than WT IL-8. The observation that the 9–50 disulfide mutant is less stable than the 7–34 mutant but has a similar dimerization constant suggests that the intrinsic stability changes on disulfide deletion do not significantly affect dimerization.
IL-8 disulfide mutants exist as molten globules in the monomeric state
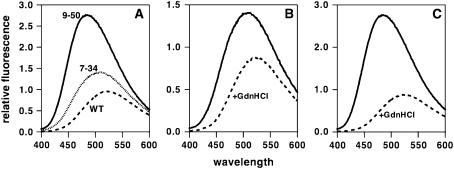
ANS is a hydrophobic fluorescent molecule that has been extensively used as a probe for the molten globule state. The molten globule has nascent structural specificity, a native-like tertiary fold, and has been proposed to represent a critical intermediate in the protein-folding process (15,16). ANS is believed to bind exposed hydrophobic patches that are characteristic of these intermediate states, resulting in a blue shift in its fluorescence maximum. Most importantly, ANS generally binds neither the fully folded nor the completely unfolded state of the protein.
The λmax of ANS fluorescence is 524 nm, and for IL-8 (1–66) monomer, λmax does not change either before or after addition of denaturant, consistent with the observation that it unfolds via a two-state mechanism from fully folded to fully unfolded, with no partially folded or molten globule intermediates (Fig. 4). For the IL-8 7–34 disulfide mutant, λmax is blue shifted by ∼10 nm, which shifts to 524 nm on addition of GdnHCl. The IL-8 9–50 disulfide mutant elicited a much larger ANS fluorescence blue shift, which also reverted to 524 nm on adding GdnHCl (Fig. 4). These observations indicate that both 7–34 and 9–50 disulfide mutants show molten globule-like characteristics, which are lost in the GdnHCl-induced unfolded state.
FIGURE 4.
ANS fluorescence spectra in presence of IL-8 monomer (dashed line), 7–34 (dotted line), and 9–50 (solid line) disulfide mutants (A). ANS fluorescence spectra of 7–34 disulfide (B) and 9–50 disulfide (C) mutants in absence (solid line) and presence (dotted line) of GdnHCl.
Structural characteristics of disulfide mutants in the dimeric state
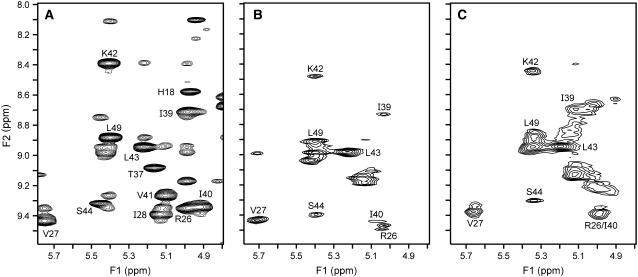
We have characterized both disulfide deletion mutants by NMR spectroscopy. The spectra of both mutants showed β-sheet chemical shifts (NH > 9 ppm; CαH > 5 ppm) and characteristic upfield-shifted peaks for Val58 Cγ2H3 and Ile22 CγH3, indicating that both mutants are folded and retain an IL-8-like fold. However, NOESY spectra for both mutants predominantly show broad and/or weak peaks. A region of the NOESY spectra showing sequential NOEs of the β-sheet amide protons is shown (Fig. 5). Compared to the WT, the cross peaks in the mutants are either broad, weak, or absent, suggesting an expanded molten globule-like structure. The NMR data do indicate relatively better defined cross peaks for the 7-34 mutant compared to the 9-50 mutant, suggesting that the former mutant is more structured, which is consistent with the observations that the 9–50 disulfide mutant is relatively less stable and shows a greater molten globule-like structure in the monomeric state.
FIGURE 5.
Sections of the 750-MHz 2D NOESY 1H NMR spectrum of the (A) WT, (B) 7–34, and (C) 9–50 disulfide mutants showing sequential nuclear Overhauser effects from β-sheet amide protons. The amide chemical shifts are labeled in all the panels.
In principle, line broadening could be attributed to chemical exchange between monomer and dimer, as ∼20% of the protein exists as a monomer for both disulfide mutants at NMR concentrations (∼0.5 mM). Chemical shifts of core residues Lys42 and Ser44 are essentially identical in both the monomer and dimer, whereas shifts of the dimer interface residue Val27 differ by >1 ppm (8). However, the NMR spectra reveal that the loss of intensity in the disulfide mutants is greater for Lys42 and Ser44 than for Val27 (Fig. 5), indicating that line broadening is not a result of monomer-dimer equilibrium but is caused by loss of core structure. NMR studies using recombinant proteins of both the monomer and dimer containing the disulfide deletions are necessary for a more quantitative description of the similarities and differences between the mutants in both the monomer and dimer structures.
Disulfides in structure, stability, and function
The importance of disulfides for protein structure and stability has been well characterized in monomeric proteins, and it has been observed that loss of structure and stability as a result of deleting disulfides in these proteins invariably results in loss of function (17,18). How disulfides influence stability and fold of noncovalent dimeric proteins and their relationship to function are less well studied. In one study, deleting disulfide in superoxide dismutase prevents dimer formation and results in marginally stable monomers (19). The chemokine IL-8 exists as both monomer and dimer, and structures of both monomeric and dimeric IL-8 are known (7,8). Structures, sequence analysis, and mutagenesis studies have shown that the conserved hydrophobic residues are packed against each other in the protein core, and H bonding and van der Waals interactions stabilize the dimer interface (20). We observe in this study that deleting disulfides results in reduced stability of the monomer, but the ability to form dimers is only marginally perturbed. This indicates that stabilizing interactions across the dimer interface in the native protein are essentially conserved in the disulfide mutants. Our observations further suggest that the stability imparted by the disulfides at the monomer level is not coupled to interactions that stabilize the dimer interface.
IL-8 function involves binding to receptors that belong to the G-protein-coupled receptor (GPCR) class and to cell surface glycosaminoglycans (GAG), and it has been shown that the monomer is the active form for receptor function, and the dimer is the active form for GAG binding (9–11). Deleting either disulfide in IL-8 results in >1000-fold loss in receptor activity, indicating that both disulfides are critical for function in the folded monomer (20). Our current studies show that the disulfide mutants are folded but adopt a molten globule-like structure, indicating that the conserved hydrophobic residues determine the protein fold, and the disulfides stabilize the structure that is essential for function. In summary, our results together indicate that individual contribution of both disulfides to global structure and stability is fundamental and essential for protein activity and has only a minimal effect on dimer formation.
Acknowledgments
We thank Dr. Wayne Bolen for access to the fluorometer, Phil Owen for peptide synthesis, and Dr. Shanmin Zhang for technical support.
This work was supported in part by grants from the National Institutes of Health AI-069152 and the John Sealy Endowment Fund.
Editor: Heinrich Roder.
References
- 1.Anfinsen, C. B., and E. Haber. 1961. Studies on the reduction and re-formation of protein disulfide bonds. J. Biol. Chem. 236:1361–1363. [PubMed] [Google Scholar]
- 2.Betz, S. F. 1993. Disulfide bonds and the stability of globular proteins. Prot. Sci. 2:1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wedemeyer, W. J., E. Welker, M. Narayan, and H. A. Scheraga. 2000. Disulfide bonds and protein folding. Biochemistry. 39:4207–4216. [DOI] [PubMed] [Google Scholar]
- 4.Pace, C. N., E. J. Hebert, K. L. Shaw, D. Schell, V. Both, D. Krajcikova, J. Sevcik, K. S. Wilson, Z. Dauter, R. W. Hartley, and G. R. Grimsley. 1998. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J. Mol. Biol. 279:271–286. [DOI] [PubMed] [Google Scholar]
- 5.Collins, E. S., J. Wirmer, K. Hirai, H. Tachibana, S. I. Segawa, C. M. Dobson, and H. Schwalbe. 2005. Characterization of disulfide-bond dynamics in non-native states of lysozyme and its disulfide deletion mutants by NMR. ChemBioChem. 6:1619–1627. [DOI] [PubMed] [Google Scholar]
- 6.Burrows, S. D., M. L. Doyle, K. P. Murphy, S. G. Franklin, J. R. White, I. Brooks, D. E. McNulty, M. O. Scott, J. R. Knutson, D. Porter, P. R. Young, and P. Hensley. 1994. Determination of the monomer-dimer equilibrium of interleukin-8 reveals it is a monomer at physiological concentrations. Biochemistry. 33:12741–12745. [DOI] [PubMed] [Google Scholar]
- 7.Clore, G. M., E. Appella, M. Yamada, K. Matsushima, and A. M. Gronenborn. 1990. Three-dimensional structure of interleukin 8 in solution. Biochemistry. 29:1689–1696. [DOI] [PubMed] [Google Scholar]
- 8.Rajarathnam, K., I. Clark-Lewis, and B. D. Sykes. 1995. 1H NMR solution structure of an active interleukin-8 monomer. Biochemistry. 34:12983–12990. [DOI] [PubMed] [Google Scholar]
- 9.Clark-Lewis, I., B. Dewald, M. Loetscher, B. Moser, and M. Baggiolini. 1994. Structural requirements for IL-8 function identified by design of analogs and CXC chemokine hybrids. J. Biol. Chem. 269:16075–16081. [PubMed] [Google Scholar]
- 10.Rajarathnam, K., B. D. Sykes, C. M. Kay, B. Dewald, T. Geiser, M. Baggiolini, and I. Clark-Lewis. 1994. Neutrophil activation by monomeric interleukin-8. Science. 264:90–92. [DOI] [PubMed] [Google Scholar]
- 11.Hoogewerf, A. J., G. S. V. Kuschert, A. Proudfoot, F. Borlat, I. Clark-Lewis, C. A. Power, and T. N. C. Wells. 1997. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 36:13570–13578. [DOI] [PubMed] [Google Scholar]
- 12.Rajarathnam, K., B. Dewald, M. Baggiolini, B. D. Sykes, and I. Clark-Lewis. 1999. Disulfide bridges in interleukin-8 probed using non-natural disulfide analogs: dissociation of roles in structure and function. Biochemistry. 38:7653–7658. [DOI] [PubMed] [Google Scholar]
- 13.Fernando, H., G. T. Nagle, and K. Rajarathnam. 2007. Thermodynamic characterization of interleukin-8 monomer binding to CXCR1 receptor N-terminal domain. FEBS J. 274:241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoro, M. M., and D. W. Bolen. 1988. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 27:8063–8068. [DOI] [PubMed] [Google Scholar]
- 15.Semisotnov, G. V., N. A. Rodionova, O. I. Razgulyaev, V. N. Uversky, A. F. Gripas, and R. I. Gilmanshin. 1991. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 31:119–128. [DOI] [PubMed] [Google Scholar]
- 16.Redfield, C., B. A. Schulman, M. A. Milhollen, P. S. Kim, and C. M. Dobson. 1999. α-Lactalbumin forms a compact molten globule in the absence of disulfide bonds. Nature Struct. Biol. 6:948–952. [DOI] [PubMed] [Google Scholar]
- 17.Pace, C. N., G. R. Grimsley, J. A. Thomson, and B. J. Barnett. 1988. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J. Biol. Chem. 263:11820–11825. [PubMed] [Google Scholar]
- 18.Vogl, T., R. Brengelmann, H.-J. Hinz, M. Scharf, M. Lötzbeyer, and J. W. Engels. 1995. Mechanism of protein stabilization by disulfide bridges: Calorimetric unfolding studies on disulfide-deficient mutants of the α-amylase inhibitor tendamistat. J. Mol. Biol. 254:481–496. [DOI] [PubMed] [Google Scholar]
- 19.Lindberg, M. J., J. Normark, A. Holmgren, and M. Oliveberg. 2004. Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc. Natl. Acad. Sci. USA. 101:15893–15898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowman, H. B., W. J. Fairbrother, P. H. Slagle, R. Kabakoff, J. Liu, S. Shire, and C. A. Hebert. 1997. Monomeric variants of IL-8: Effects of side chain substitutions and solution conditions upon dimer formation. Protein Sci. 6:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]