Abstract
A novel method is proposed using nanometer-sized diamond particles as detection probes for biolabeling. The advantages of nanodiamond's unique properties were demonstrated in its biocompatibility, nontoxicity, easily detected Raman signal, and intrinsic fluorescence from its natural defects without complicated pretreatments. Carboxylated nanodiamond's (cND's) penetration ability, noncytotoxicity, and visualization of cND-cell interactions are demonstrated on A549 human lung epithelial cells. Protein-targeted cell interaction visualization was demonstrated with cND-lysozyme complex interaction with bacteria Escherichia coli. It is shown that the developed biomolecule-cND complex preserves the original functions of the test protein. The easily detected natural fluorescent and Raman intrinsic signals, penetration ability, and low cytotoxicity of cNDs render them promising agents in multiple medical applications.
INTRODUCTION
Carbon exists in many forms that provide various applications in the fields of electronics and biotechnology. Carbon paste, graphite, carbon fibers, porous carbon, and glassy carbon are used for biosensors; other newly developed promising carbon derivatives such as carbon nanotubes, fullerenes, and diamond nanofilms have all found their ways to applications in nanotechnology (1–4). Among the various forms of carbon, diamond is by itself unique and usually recognized by its exceptional hardness, thermal properties, excellent optical properties, and electrochemical inertness. In addition, from the point of view of bioapplications, available with variable sizes in nanoscale, diamond surfaces provide a convenient platform for bioconjugation (2,5–10). The hydrogen-carbon bonding characteristics of most diamond surfaces are stable (11) and convenient for bioconjugation either chemically (covalently or noncovalently) or physically (adsorption). Recently, the physical and chemical properties of nanometer-sized diamond particles have attracted great attention for their promising applications in the field of nanobiotechnology (12,13). The methods of functionalization and modification of different nanoparticles are developing (14); recently some work has been done concerning surface functionalization (15) and modification of diamond film surfaces with biomolecules such as DNA (5–7); different proteins, e.g., interacting antigens and antibodies (10,12), fluorescent dye–labeled proteins (16), etc., have been immobilized on the diamond thin films. In this respect, nanometer-sized diamond particles allow easier biomolecule immobilization while at the same time they possess reproducible electrochemical behavior and useful physical characteristics (8,17,18).
These promising applications also pose great public concern on the safety of these nanoparticles (19–24). The problem of cytotoxicity is directly related to the common problem of health effects of nanomaterials, and direct investigations are necessary to understand the mechanisms of nanotoxicity. On the other hand, nanoparticles have potential as nanosized probes and sensors for tissues (e.g., intravascular probes; probes for tumor targeting) and directly for cells for both diagnostic (e.g., imaging) and therapeutic purposes (e.g., drug delivery) (25,26). Therefore, for any sensible application in a biosystem, their nontoxicity and biocompatibility are necessary conditions; easy surface functionalization and modification as well as detection are crucial for nanoparticle biosensing. It is hoped the desired nanoparticle-biomolecule complexes can interact specifically or nonspecifically with components of investigated bio-objects and that, as a result, many interactions can be studied in detail.
The interaction of the nanodiamonds and cells can be studied using spectroscopic methods, in particular Raman spectroscopy. Raman scattering can be excited at any wavelength that does not photobleach the investigated system. It has high spectral and spatial resolution, so that spectral overlapping can be largely avoided. However, Raman signal intensity is usually low, and the spectra can be complex for many nanoparticles and biomolecules, so the few works using Raman detection for biosensing have been concentrated mainly on surface-enhanced Raman scattering (SERS) detection (27). Metal (Ag) nanoparticles covered with different Raman-active dyes were used to label the investigated system. Unfortunately, dye labeling poses uncertain toxicity to the biomolecules or cells that are under investigation. To avoid this complication, nanodiamonds are promising candidates. Raman investigation is a noninvasive method and can be performed in ambient conditions. We show that nanodiamond particles can label the cells. The typical Raman spectrum of diamonds exhibits a sharp peak located at 1332 cm−1 for phonon mode of the sp3 bonding carbons (28). It is quite intense and narrow and is not affected by the surface functional groups or the connected biomolecules of interest. This peak is usually accompanied by two graphite modes caused by graphitic structures (the D- and G-bands of sp2 carbons at 1355 cm−1 and 1575 cm−1, respectively) on the surfaces. However, the graphitic structure usually can be removed during the carboxylation stage, as a result of strong acid washing, when functional groups are generated on the diamond surface. Therefore, this diamond peak is isolated, and the Raman absorption cross section is large enough so this peak can be used as an indicator for the location of diamond when the laser scans spatially across the sample. However, when the diamond size decreases, significant surface atoms dominate the particle, so the 1332 cm−1 peak also decreases. In our size-dependent Raman analysis, diamond particle sizes of 50 nm and above could provide a 1332 cm−1 peak clear enough for Raman detection. For smaller diamonds, one suffers the competition of D- and G-band intensities. Nevertheless, it is still possible to use the broader D- and G-bands for detection.
In this article, the possibility of using nanometer-sized diamond particles as nanobioprobe is presented. The biocompatibility is demonstrated by the cytotoxicity test of carboxylated nanometer-sized diamonds (cNDs, average diameters 5 and 100 nm) in A549 human lung epithelial cells. To emphasize its uniqueness, we present the interaction of cNDs with cells; the undamaging penetration of cNDs into cells and their accumulation in cells; and the distribution of cND within cells as visualized by detection of nanodiamond spectroscopic signals (Raman and fluorescence). The principles of using nanodiamonds for cell labeling and the microscopic and spectroscopic methods of detection are also discussed. The connection of diamonds and proteins is determined by the functional groups created on the diamond surfaces through surface carboxylation. Carboxylated nanodiamond conjugated with test protein, lysozyme, through physical adsorption is considered as a model nanobioprobe, and its interaction with bacteria Escherichia coli is analyzed. The developed nanobioprobe allows visualization of the protein-bacteria interaction via the detection of diamond Raman signal and optical imaging while preserving the original functions of the test protein.
MATERIALS AND METHODS
Nanodiamond functionalization
Two different sizes of nanodiamonds, 5 nm (5-ND, Microdiamant AG, Lengwil, Switzerland) and 100 nm (100-ND, General Electric, Fairfield, CT), were used. The 100-ND has a mean particle size of 110 nm with standard deviation 25 nm in size when purchased. The 5-ND has a very uniform mean size of 5 nm and sharp distribution (∼4–6 nm) as seen from our scanning electron microscope observation (SEM JEOL JSM6500F, Japan). Both nanodiamond sizes have variable shapes. SEM images were also obtained to visualize the surface morphology of the carboxylated NDs. The carboxylation of NDs, forming surface functional COOH groups, followed standard chemical methods (5,17,18) consisting of sample heating in a 9:1 mixture of concentrated H2SO4 and HNO3 at 75°C for 3 days, then in 0.1 M NaOH aqueous solution at 90°C for 2 h, finally in 0.1 M HCl aqueous solution at 90°C for 2 h, extensive rinsing with deionized water, separation by sedimentation with a centrifuge at 12,000 rpm, and drying. After the carboxylation, the prepared cNDs were examined using Fourier transform infrared spectrometer (ABB Bomem FTIR MB154, Zurich, Switzerland) with samples in vacuum chamber at 10−3–10−5 torr range or in ambient conditions (17). Carboxylation is characterized by well-resolved lines of C=O stretching and O-H bending of carboxyl groups on the nanodiamond surface in the ranges 1720–1780 and 1630–1640 cm−1, respectively. For FTIR measurements, the water suspensions of NDs were dropped on Si substrate and dried. No special measures were taken to prevent suspensions from aggregation, but concentrations were varied in wide range in this experiment, so the degree of aggregation also should vary. We did not observe the dependences of optical properties on the degree of aggregation of nanodiamonds.
The A549 cell sample preparation
A549 human lung adenocarcinoma cells were maintained in RPMI-1640 medium (Invitrogen, Carlsbad, CA). These cells were cultured at 37°C and 5% CO2 in a humidified incubator (310/Thermo, Forma Scientific, Marietta, OH). The A549 cells were treated with 0–1000 μg/ml 5-cNDs or 100-cNDs for 4 h, then washed with isotonic phosphate-buffered saline (PBS, pH 7.4) and recultured in complete RPMI-1640 medium for 20–24 h. The cells' survival rate was measured by Cytotoxicity MTT Assay. For the fluorescent investigation and Raman mapping investigation, the A549 cells were cultured analogously but on coverslips or Si substrates, correspondingly.
The cytotoxicity test
The cells were plated in 96-well plates at a density of 1 × 104 cells/well for 16–20 h and then treated with various concentrations of 5-cND or 100-cND for 4 h in serum-free RPMI-1640 medium. After the treatment, the cells were washed twice with PBS and were recultured in complete RPMI-1640 medium (containing 10% PBS) for 2 d. Subsequently, the medium was replaced, and the cells were incubated with 0.5 mg/ml of MTT in complete RPMI-1640 medium for 4 h. The surviving cells converted MTT to formazan, which generates a blue-purple color when dissolved in dimethyl sulfoxide. The intensity was measured at 565 nm using a plate reader (OPTImax; GE Healthcare, Little Chalfont, UK) for enzyme-linked immunosorbent assays. The relative percentage of surviving cells was calculated by dividing the absorbance of treated cells by that of the control in each experiment.
Fluorescence investigation
To examine the location of nanodiamonds in lung epithelial cells, the A549 cells were cultured on a coverslip, which was kept in a 60-mm petri dish for 16–20 h before treatment, then treated with 100-nm cNDs (100 μg/ml, 4 h), washed twice with isotonic PBS (pH 7.4), and were cultured in complete RPMI medium (containing 10% serum) for 20 h. The samples were fixed in ice-cold 4% paraformaldehyde solution in PBS for 1 h at 37°C, washed three times with PBS, and cell nonspecific binding sites were blocked in PBS containing 10% normal bovine serum and 0.25% Triton X-100 for 1 h. The cytosol was stained with 1:50 Cy3-labeled mouse anti-β-tubulin (Sigma, St. Louis, MO) for 30 min. The coverslips were washed three times with 0.25% Triton X-100 in PBS. The nuclei were stained with 2.5 μg/ml Hoechst 33258 (Sigma, St. Louis, MO) for 30 min. The coverslips were washed three times with 0.25% Triton X-100 in PBS. Finally, the samples were stored in the dark at 4°C until examined under a Leica confocal laser-scanning microscope (TCS SP2, Mannheim, Germany) that was equipped with a UV laser (351/364 nm), an argon laser (457/488/514 nm), and a HeNe laser (543 nm/633 nm).
Quantum dots (QD), for comparison and estimation of cND fluorescence, CdSe/ZnS QD 525 (Catalog number: 1104-1, designated QD-IgG) were purchased from Quantum Dot (Hayward, CA). The fluorescence spectra of cND and QD were measured with an α-SNOM (Witec, Ulm, Germany) confocal Raman spectrometer with excitation at 488 nm using an ion laser (Melles Griot, Carlsbad, CA).
The Raman spectroscopic measurements and Raman mapping
The α-SNOM confocal Raman spectrometer equipped with an x-y scanning stage, with 488-nm wavelength laser excitation, was used. To avoid laser damage, low laser power (<1 mW in focal spot) was employed. The distribution of diamond Raman signal (1332 cm−1) across the sample's studied area was mapped. The prepared Si wafers with the cell samples were washed three times with PBS and stored in the dark at 4°C until measurements. The water immersion objective (65×) was used for direct measurements of the cells in the medium.
The lysozyme adsorption on carboxylated nanodiamond
The protein lysozyme (Amresco, Solon, OH) was attached to 100-cND via physical adsorption according the method described before (17). In brief, lysozyme in concentration 180–200 μM was dissolved in PBS (pH 6.5–7). The protein concentration was checked with UV/Visible spectrometer (Jasco V-550, Jasco, Tokyo, Japan) using the solution adsorption at 280 nm (29). A molar absorbance of lysozyme at 280 nm (3.7547 × 104 M−1 cm−1) served to calibrate the protein concentration by the measured absorbance at the Soret band maximum. The initial concentration of lysozyme in solution was measured before adsorption; then cND was added to the solution in concentration 4–10 mg/ml. To ensure equilibration of the adsorption, the protein solution and the cND powder were thoroughly mixed together with a shaker for 2 h, after which the mixture was centrifuged several times and washed with deionized water. After separation of cND with adsorbed lysozyme, the residual concentration of protein in the supernatant was measured. The amount of lysozyme on the cND surface was estimated by the difference between initial and residual protein concentrations in solution. The maximum amount of lysozyme that can be adsorbed by 1 mg of 100 nm carboxylated nanodiamond was calculated to be 80 ± 10 μg; which corresponds to a few thousand (∼3700) lysozyme molecules on one nanodiamond surface, neglecting the aggregation of the nanodiamonds. The as-prepared nanodiamond-biomolecule complexes were analyzed with FTIR spectra. The cND-lysozyme complex is stable and useful for biomedical applications (17).
Interaction of cND-protein complex with E. coli
Bacteria E. coli were cultivated in Luria-Bertani (LB) medium in a shaking incubator at 37°C. The optical density level at wavelength 600 nm (OD600) should reach 0.5 when the samples were analyzed with a UV/Visible spectrometer. After incubation the bacteria were separated by centrifugation at 12,000 rpm for 5 min and resuspended in bidistilled water or in PBS solution for spectroscopic measurements. The E. coli water suspension was mixed together with cND or with lysozyme-cND complex suspensions. The mixtures were stirred for 30–200 s and then dropped on Si substrate and rapidly dried. The Raman spectra were measured with the α-SNOM confocal Raman spectrometer. For the survival test, 25 μl of E. coli suspension in PBS was mixed with 50 μl suspension of 100-cND–lysozyme complexes in PBS, and the cND concentration was ∼1 mg/ml; corresponding average concentration of lysozyme estimated in accordance with UV/Visible absorption spectroscopic measurements was ∼20–70 μM. For comparison, E. coli suspension was mixed with PBS and 50 μM lysozyme solution in PBS. The mixtures were incubated for 60-90 min at 37°C. Then the bacterial suspensions were diluted 104-fold, thoroughly stirred, and spread in 20 μl on agar gel in petri dishes. The petri dishes were incubated at 37°C for 12–16 h; then the bacterial colonies in every dish were counted.
RESULTS
Cytotoxicity test
In cytotoxicity experiments, we specifically used cND, as these would be the building blocks for further connection to biomolecules of interest. Fig. 1 a is a typical view from conventional optical microscope for A549 cells treated with 1 mg/ml of 100-nm cND. Fig. 1 b is the cell survival rate as measured by MTT assay for both 5-nm cND and 100-nm cND–treated A549 cells. As depicted in Fig. 1, both 5-nm and 100-nm cNDs did not significantly induce the cell death in A549 cells. Interestingly, we found these nanodiamonds that located on A549 cells (Fig. 1 a, and see Fig. 4 a) via a concentration-dependent manner (data not shown). In the cytotoxicity test, because a high concentration of cND was used (1 mg/ml), we observed the aggregation of cNDs on the cells. The aggregation can be so large that cND aggregates are visible in an optical microscope.
FIGURE 1.
Cytotoxicity test of cNDs on A549 human lung epithelial cell. (a) A typical view from conventional optical microscope for A549 cells treated with 1 mg/ml of 100-nm cND. (b) The cell survival rate as measured by MTT assay for both 5-nm cND and 100-nm cND–treated A549 cells.
FIGURE 4.
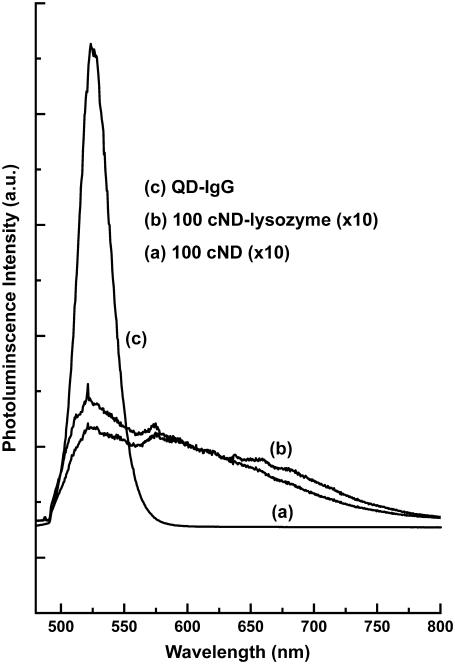
Comparison of the photoluminescence spectra of the QD and cND. The spectra are obtained using 488-nm wavelength laser excitation.
Raman spectroscopic detection
For practical applications, it is desired that only a few cND nanoparticles would be used when visualization could not be obtained from a conventional optical microscope. Fig. 2 a displays a typical SEM image of the 100-nm cNDs directly deposited on a silicon wafer. Fig. 2 b is the typical Raman 1332 cm−1 signal of sp3 carbons of the nanodiamonds from Fig. 2 a. When the laser scans with a step 0.5 μm along a 30-μm line as indicated in Fig. 2 c of a 100-nm cND-with-A549 cell system, and the intensity distribution of the 1332 cm−1 peak is plotted against the distance, the location of the aggregated cNDs can be found from the plot in Fig. 2 d. In Fig. 3, confocal Raman mapping was performed on a 100-nm cND–A549 cell system. In Fig. 3, a and c, the concentration of cND used was 10 μg/ml to interact with the cell, whereas in Fig. 3, b and d, the concentration of cND was 1 μg/ml. In each set of figures, the optical images of two A549 cells and the xy-Raman mapping along the square box indicated in Fig. 3, a and b, are shown. In Fig. 3 d, a single cND or cND aggregate can be located using this mapping technique.
FIGURE 2.
Raman scanning on A549 human lung epithelial cell. (a) A typical scanning electron microscope image of 100-nm cND with the diamonds directly deposited on a single crystal silicon wafer. (b) Raman spectrum of 100-nm diamond. (c) Optical image of an A549 cell interacted with 100-nm cNDs. The sharp Raman signature in c can be used as an indicator to locate the diamond position in the cells. (d) The diamond Raman peak intensity distribution versus distance across the line indicated in c, scanning with a step of 0.5 μm.
FIGURE 3.
Confocal Raman mapping of a single cND aggregation in A549 cell line. (a and b) Two A549 cells can be seen under optical microscope (water immersion objective, 65×) with cND concentration 10 μg/ml (a) and 1 μg/ml (b), respectively; (c and d) corresponding confocal Raman mapping locked on the diamond signals reveals the positions of the cND.
Confocal fluorescence microscopic investigation
There have been many applications using commercial semiconductor nanocrystals (quantum dots, QDs) for their well-defined spectral characteristics (25). Here we provide an alternative to use the nontoxic, easy functionalized/conjugated cNDs for biolabeling. In Fig. 4, a comparison of the fluorescence (excited with a 488-nm wavelength laser) of commercial quantum dots (QD-IgG) versus cND/protein-attached cNDs is presented. A broad fluorescence with maximum intensity centered at ∼525 nm was observed for 100-cND and 100-cND–lysozyme. The ratio of fluorescence intensity of QD-IgG/cND-lysozyme was estimated to be ∼30. It has been reported that high-energy treatment of ND creates more defect centers, and the fluorescence can be enhanced by two orders of magnitude, which make it closer to most QDs (30).
During the interaction of nanodiamonds with A549 cells, it appears that the diamond particles aggregated in the area of the cell's cytoplasm. A second spectroscopic method was applied to show the exact location of the cND within the cell, which can be identified using the natural fluorescence of nanodiamonds. We demonstrate the confocal fluorescent images in Figs. 5–7. In Fig. 5, the cytoskeleton and nuclei were stained with the Cy3-labeled anti-β-tubulin and Hoechst 33258, respectively (Fig. 5, a and b). In Fig. 5, c and d, intensive signal exhibited by 100-cNDs was collected in the same cells (at excitation wavelength 488 nm the emitted light was collected at wavelengths 500–530 nm; with excitation at 633 nm, the emitted signal was detected in the range 640–720 nm). When the images of Fig. 5, a–d, were merged, the results in Fig. 5 e demonstrate that the Cy3 dye can be replaced by 100 cNDs in our experiments. Emission at different colors has been reported previously from various defect centers in nanodiamonds (31). Through the confocal fluorescence image mapping, we confirm the coexistence of the nanodiamond particles with the A549 cells. These observed images of cND presumably could also be caused by some scattering, as the excitation wavelength is very close to detection range. In separate experiments, we have obtained the cND fluorescence spectra using various wavelengths of laser excitation, and at 488 nm excitation, the emission is maximum in ranges 500–530 nm and 580–680 nm (Fig. 4); at 532 nm excitation the emission is observed in range 580–700 nm, and at 633 nm, in the range 640–720 nm (results not shown) (32).
FIGURE 5.
Confocal fluorescence images of an A549 cell and carboxylated 100 nm diamond. (a) The cell nuclei were dyed with Hoechst 33258 to reveal the position of the nucleus. (b) The cell tissue was dyed with anti-β-tubulin (Cy3) to reflect the cytoskeleton of the cells. (c) The cells were interacted with 100-nm cNDs and excited with 488-nm wavelength, and the emission was collected in the range of 500–530 nm. (d) Same as in c but exciting wavelength was 633 nm and emission was collected in the range 640–720 nm. (e) Merging the images of a–d.
FIGURE 6.
Cross-sectional scan on a single A549 cell. (a) A series of confocal fluorescence images at changing position in the z direction from the top (upper left) down (bottom right) in one of the A549 cells in Fig. 3. When the confocal microscope is scanned in the vertical direction with steps 1 μm from top to bottom, the distribution of the anti-β-tubulin surrounding the nucleus can be clearly visible. (b) The fluorescence images reveal that diamond can penetrate into the cell and aggregate in cytoplasm. At this concentration of diamond (100 μg/ml), the cells survive the cytotoxicity test. The observed light signal from the 100-cNDs was excited with wavelength 488 nm, and the emission was collected in the range of 500–530 nm. (c) A phase-contrast image of the cell. (d) The merging of all three types of images.
FIGURE 7.
Cross-sectional scan on a single A549 cell. A series of confocal fluorescence images at changing position in the z direction with 1-μm steps from the top (upper left) down (bottom right) in one of the A549 cells in Fig. 3. The position of a single (or aggregated) cND can be clearly visible (shown in green). At this concentration of diamond (1 μg/ml), the observed signal from the 100-cNDs was excited with wavelength 488 nm, and the emission was collected in the range of 500–530 nm.
To further trace the interaction of cNDs and the cell, a 3-D scan was performed with a confocal fluorescence microscope. With excitation at 488 nm and emission collected in the range of 500–530 nm, a series of images of Cy3-labeled anti-β-tubulin and cND in different positions on the z-axis are plotted in Fig. 6, a and b. When the confocal microscope was scanned in the vertical direction with 1-μm steps from top to bottom, the distribution of the anti-β-tubulin surrounding the nucleus can be clearly visible. Corresponding phase-contrast optical microscope images are also plotted for reference (Fig. 6 c). When cNDs penetrated inside the cells, they indeed resided near the cytoplasm as revealed by different z-position fluorescent images. In Fig. 6 a, the A549 cell has been dyed with fluorescent anti-β-tubulin to reveal the image of the cellular cytoskeleton. In Fig. 6 b, the collected light signal from the cNDs is plotted. The fluorescent images completely traced the contour of the β-tubulin around the outside rim of the nucleus. Next, we further show that the naturally fluorescent cND provide fluorescence signals strong enough for observation even at low concentration. In Fig. 7, the same experiments as in Figs. 5 and 6 were performed, except the cND concentration was 1 μg/ml. We located one or two cND and cND aggregates within the cell as revealed by a series of confocal fluorescence images in the z direction.
Interaction of cND-protein complex with E. coli
Fig. 8 a plots the survival rate of the growth of E. coli colonies after 30 min of treatment with two different cND-lysozyme complexes and after incubation. The diagram averages the data of eight independent experiments. One can see that after the treatment with cND-lysozyme complex, the number of colonies decreases significantly relative to the control, so the cND-lysozyme complex displays antibacterial activity, equivalent to high activity of pure lysozyme in solution within experimental error. This demonstrates that the adsorption of lysozyme on cNDs practically does not significantly alter the original functionality of the protein for the case of lysozyme. Further, the interaction of the lysozyme with bacteria E. coli could be visualized via the detection of the Raman intrinsic signal of the 1332 cm−1 peak as it was in the cND-cell interaction demonstrated in Figs. 2 and 3. In that case, E. coli were treated with the cND-lysozyme complex, and the interaction was first observed with an optical microscope (objective 100×) and then by a confocal Raman spectrometer. In Fig. 8 b, optical images of the E. coli can be clearly seen, but not the reacting cND-lysozyme complex. In the Raman mapping, the diamond spectral signal was locked, and an x-y scan was performed across a 10 μm × 10 μm area with 0.2-μm step, and the diamond signal was plotted.
FIGURE 8.
Raman mapping of the cND-lysozyme interaction with E. coli. (a) Standardized number of E. coli colonies after 30 min of treatment with (I) PBS control; (II) lysozyme solution; (III) suspension of lysozyme-5cND conjugates, and (IV) suspension of lysozyme-100-cND conjugates. The measurements were obtained from eight independent experiments, and error bars were drawn using standard deviation. (b) The interaction of E. coli with cND-lysozyme complex as viewed with conventional optical microscope (objective 100×) and confocal Raman spectrometer. In the optical image, E. coli can be seen. The Raman signal of the diamond peak (1332 cm−1) was locked and scanned across a 10 μm × 10 μm area, and the distribution of diamond signal intensity was plotted. The location of the nanodiamond is indicated in yellow.
DISCUSSION
Cytotoxicity of nanodiamond
The cytotoxicity results indicate that even at high doses of cNDs (up to 1000 μg/ml), the cells survive even though some of the cNDs may have penetrated into the cells. This result agrees with a recent research (33) that described in detail the cytotoxcity of ND, 2–10 nm diameter, with different cell models; and the cytotoxicity test was conducted at concentrations ranging between 5 and 100 μg/ml. Our work focused on 5- and 100-nm nanodiamonds, and the cytotoxicity test was carried out at 0-1000 μg/ml. As we learned from our previous studies, nanodiamonds of 5-nm diameter, where >15% of the carbon atoms are the surface atoms, have significantly different surface properties than the larger nanodiamonds (such as 100-nm ND) (18). For applications using spectroscopic techniques (Raman or fluorescence), the choice of sizes is then essential. The nanodiamonds appear to attach to the cells even after repeated washing with PBS. The interaction mechanism of the cNDs with the cell is not clear, possibly because of the interaction of the anionic groups (−COO−) from cNDs and the positively charged groups on cell membrane outer surface (8), and further through endocytosis or phagocytosis uptake into the cells; the penetration of cND inside cells and localization in cytoplasm were further demonstrated in the following Raman and fluorescence experiments.
Raman and fluorescence mapping
In our Raman mapping investigations, the size of the separated ND particle is 100 nm; it is smaller than the diffraction limit of the optical microscope as well as the step of the scanning performed. So the mapping may not reveal the size of the aggregates; but at such a high concentration of nanodiamonds, and with no precaution taken to prevent the aggregation, the observed images in Fig. 2, c and d, should come from the aggregated ND particles. The advantage here is to neglect the perhaps complex spectroscopic information of biomolecules when conjugated with the cNDs. Raman spectral mapping (in the x,y directions) allows us to map out the spatial distribution of the complex as shown in Fig. 3. This is a feasible approach in the mapping of biomolecule interaction provided the cNDs are interacting with the biomolecules of interest. In our trial experiment the size of an A549 cell is 10–30 μm; with confocal Raman ability, the spatial resolution can easily be pushed further into the nanometer range, being limited by laser's focal spot size and estimated to be ∼300 nm. More importantly, this method is noninvasive to the cell system and can be performed in vivo. The particles are inside the cell, but to confirm this we have done the measurements with confocal fluorescent microscopy.
It has been shown that a single nitrogen-vacancy (N-V) color center in a diamond can emit light efficiently, and very remarkable photostability has been achieved (34,35). On 514-nm wavelength excitation, a single N-V center could emit intensive red fluorescence (34,36,37). In the synthetic diamonds, N-V centers as well as other emitting photoluminescence defects exist arising from the process of production (38). For the 5-nm size diamond produced using the detonation method, nitrogen and oxygen defects or impurities exist inside the grains (39). Because considerable parts of the atoms are located near the grain surface, the nanomaterial properties can differ from those of natural bulk diamonds (40). In our experiment (Figs. 4–7), we observed fluorescence arising from the natural defects/impurities of nanodiamonds without complicated pretreatment (high-energy proton and high-temperature treatments (30,37)) as a result of structural or surface defects, and a more detailed investigation of the natural fluorescence of nanodiamonds is reported elsewhere (32). Despite the weakness of the cNDs' fluorescence as compared to that of the QDs (shown in Fig. 4), we mainly focus on the “untreated” cNDs and argue that the fluorescence is strong enough for confocal fluorescence measurements. In addition, in a separate experiment, we demonstrated that the fluorescence is strong enough for the flow cytometry applications (results not shown). These results indicate that the fluorescence arising from natural defects/impurities of nanodiamonds could serve as fluorescent probes for the nanodiamonds interacting with cells under laser excitation. The fluorescence of cNDs combined with their excellent biocompatibility as demonstrated in Fig. 1, and together with the superb properties of photostability and absence of photobleaching (30,34), allow them to serve as a marker for labeling the cell in addition to the regular dye molecules. For cell labeling as illustrated in Fig. 5, the concentration of cNDs was relatively high, and the collective intensity can be strong enough because of the defects for confocal microspectroscopic detection.
In Fig. 7, the same experiments as in Figs. 5 and 6 were performed, except the cND concentration was 1 μg/ml. We located one or two cNDs (or cND aggregates) within the cell as revealed by a series of confocal fluorescence images in the z direction. These results indicate that 100 cNDs penetrated inside the cell and located in the cell cytoplasm of A549 cells, and the naturally fluorescent cND can be used as a probe for detection. In fact, it has also been shown that NDs can be ingested into the human kidney cells (30). However, the mechanism of the cNDs' penetration into cell is not clear and needs further investigation.
Monitoring protein interaction with bacteria
The methods demonstrated here enable us to visualize the biomolecule interaction if a functional molecule (proteins, DNA, etc.) specifically binds the cNDs to allow interaction with the target (e.g., cell). Therefore, to develop a bioprobe on the base of nanodiamonds, cND should be conjugated with biomolecules for specific or nonspecific interaction with a bio-object of investigation, and hence, many interactions can be studied in detail. In our previous study, the lysozyme was attached to a carboxylated nanodiamond via physical adsorption provided by electrostatic attraction between the surface-terminated anionic groups (−COO−) and the positively charged amino groups (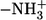 ) in the protein. The as-prepared nanodiamond-biomolecule complexes are stable and useful for biomedical applications (17). To analyze how the interaction with carboxylated nanodiamond surface affects the lysozyme's functional activity, the bacteria E. coli were treated with the cND-lysozyme complex. We concentrated on the physical adsorption in this study; actually, both carboxylated and noncarboxylated nanodiamond can adsorb organic molecules. We suppose that adsorption on surface defects, such as pores, mainly occurs for noncarboxylated nanodiamonds. Carboxylation includes strong acid treatment, so the nondiamond surface structures can be destroyed, and defect number decreases. At the same time functional groups form, which are charged and can interact with opposite charged groups of adsorbed molecules.
) in the protein. The as-prepared nanodiamond-biomolecule complexes are stable and useful for biomedical applications (17). To analyze how the interaction with carboxylated nanodiamond surface affects the lysozyme's functional activity, the bacteria E. coli were treated with the cND-lysozyme complex. We concentrated on the physical adsorption in this study; actually, both carboxylated and noncarboxylated nanodiamond can adsorb organic molecules. We suppose that adsorption on surface defects, such as pores, mainly occurs for noncarboxylated nanodiamonds. Carboxylation includes strong acid treatment, so the nondiamond surface structures can be destroyed, and defect number decreases. At the same time functional groups form, which are charged and can interact with opposite charged groups of adsorbed molecules.
The results demonstrated Raman signal from the cND-lysozyme interacting with bacteria could be detected and used as a marker to locate the position of the interacting protein lysozyme with E. coli. Note that the longer time of interaction of the cND-lysozyme and E. coli resulted in the death of the bacteria, which can be seen even under the optical microscope. In our experiment, the interaction was only 30 s. In Fig. 8 b, the diamond Raman signals (in yellow) indicate the interaction sites. The arrow in the optical image indicates an E. coli possibly destroyed by lysozyme action. Note, we ruled out the possibility that the detected signal in Fig. 8 b was simply caused by the cND and lysozyme mixture. In the preparation of the cND-lysozyme complex, high-frequency centrifugation allows separation of nonconnected lysozyme and cND. The advantage of this developed method also allows in situ (in vivo) observation of the interaction of the lysozyme with bacteria.
CONCLUSION
In conclusion, we demonstrated that nanometer-sized diamonds could be a useful probe for detecting the interaction of nanoparticles and bio-objects such as cells and bacteria. The functionalization of the surfaces and biomolecules immobilization is easy, the toxicity is low, and nanodiamond is perfectly biocompatible with live cells. The naturally embedded defects create fluorescent centers and render them possible markers for detection without additional high-energy treatment on the diamond. This can be realized for use in fluorescent imaging as a result of recently available high-sensitivity instrumentation. The intrinsic Raman peak from sp3 carbon bonding also provides a signal for detection using noninvasive Raman spectroscopic methods. The fact that the cNDs penetrate inside the cell may have promising biomedical applications.
Acknowledgments
The authors appreciate the financial support of this research by the National Science Council of Taiwan, Republic of China, under grant No. NSC-95-2120-M-259-003.
Editor: Petra Schwille.
References
- 1.Poh, W. C., K. P. Loh, W. D. Zhang, S. Triparthy, J. S. Ye, and F. S. Sheu. 2004. Biosensing properties of diamond and carbon nanotubes. Langmuir. 20:5484–5492. [DOI] [PubMed] [Google Scholar]
- 2.Carlise, J. A. 2004. Diamond films: Precious biosensors. Nat. Mater. 3:668–669. [DOI] [PubMed] [Google Scholar]
- 3.Wang, J. 2005. Carbon-nanotube based electrochemical biosensors: a review. Electroanalysis. 17:7–14. [Google Scholar]
- 4.Sotiropoulou, S., V. Gavalas, V. Vamvakaki, and N. A. Chaniotakis. 2003. Novel carbon materials in biosensor systems. Biosens. Bioelectron. 18:211–215. [DOI] [PubMed] [Google Scholar]
- 5.Ushizawa, K., Y. Sato, T. Mitsumory, T. Machinami, T. Ueda, and T. Ando. 2002. Covalent immobilization of DNA on diamond and its verification by diffuse reflectance infrared spectroscopy. Chem. Phys. Lett. 351:105–108. [Google Scholar]
- 6.Yang, W., O. Auciello, J. E. Butler, W. Cai, J. A. Carlisle, J. Gerbi, E. D. M. Gruen, T. Knickerbocker, T. L. Lasserter, J. N. Russell, L. M. Smith, and R. J. Hamers. 2002. DNA-modified nanocrystalline diamond thin-films as stable, biologically active substrates. Nat. Mater. 1:253–257. [DOI] [PubMed] [Google Scholar]
- 7.Knickerbocker, T., T. Strother, M. P. Schwartz, J. N. Russell, J. S. Butler. Jr., and R. J. Hamers. 2003. DNA-modified diamond surfaces. Langmuir. 19:1938–1942. [Google Scholar]
- 8.Huang, L. C. L., and H.-C. Chang. 2004. Adsorption and Immobilization of Cytochrome c on Nanodiamonds. Langmuir. 20:5879–5884. [PubMed] [Google Scholar]
- 9.Kong, X. L., L. C. L. Huang, C.-M. Hsu, W.-H. Chen, C.-C. Han, and H.-C. Chang. 2005. High-affinity capture of proteins by diamond nanoparticles for mass spectrometric analysis. Anal. Chem. 77:259–265. [DOI] [PubMed] [Google Scholar]
- 10.Huang, T. S., Y. Tzeng, Y. K. Liu, Y. C. Chen, K. R. Walker, R. Guntupalli, and C. Liu. 2004. Immobilization of antibodies and bacterial binding on nanodiamond and carbon nanotubes for biosensor applications. Diamond Relat. Mater. 13:1098–1102. [Google Scholar]
- 11.Cheng, C.-L., C.-F. Chen, W.-C. Shaio, D.-S. Tsai, and K.-H. Chen. 2005. The CH stretching features on diamonds of different origins. Diamond Relat. Mater. 14:1455–1462. [Google Scholar]
- 12.Kossovsky, N., A. Gelman, H. J. Hnatyszyn, A. Rajguru, R. L. Garrell, S. Torbati, S. S. F. Freitas, and G.-M. Chow. 1995. Surface-modified diamond nanoparticles as antigen delivery vehicles. Bioconjugate Chem. 6:507–511. [DOI] [PubMed] [Google Scholar]
- 13.Puzyr', A. P., I. O. Pozdniakova, and V. S. Bondar'. Design of a luminescent biochip with nanodiamonds and bacterial luciferase. 2004. Phys. Solid State. 46:761–763. [Google Scholar]
- 14.Katz, E., and I. Willner. 2004. Integrated nanoparticle-biomolecule hybrid systems: synthesis, properties, and applications. Angew. Chem. Int. Ed. Engl. 43:6042–6108. [DOI] [PubMed] [Google Scholar]
- 15.Krüger, A., F. Kataoka, M. Ozawa, T. Fujino, Y. Suzuki, A. E. Aleksenskii, A. Ya. Vul', and E. Osawa. 2005. Unusually tight aggregation in detonation diamond: identification and disintegration. Carbon. 43:1722–1730. [Google Scholar]
- 16.Clare, T. L., B. H. Clare, B. M. Nichols, N. L. Abbott, and R. J. Hamers. 2005. Functional monolayers for improved resistance to protein adsorption: oligo(ethylene glycol)-modified silicon and diamond surfaces. Langmuir. 21:6344–6355. [DOI] [PubMed] [Google Scholar]
- 17.Chung, P.-H., E. Perevedentseva, J.-S. Tu, C. C. Chang, and C.-L. Cheng. 2006. Spectroscopic study of bio-functionalized nanodiamonds. Diamond Relat. Mater. 15:622–625. [Google Scholar]
- 18.Tu, J.-S., E. Perevedentseva, P.-H. Chung, and C.-L. Cheng. 2006. Size-dependent surface CO stretching frequency investigations on nanodiamond particles. J. Chem. Phys. 125:174713–174717. [DOI] [PubMed] [Google Scholar]
- 19.Warheit, D. 2004. Nanoparticles: health impact. Mater. Today. 7:32–35. [Google Scholar]
- 20.Colvin, V. 2003. The potential environmental impact of engineered nanomaterials. Nature Biotech. 21:1166–1170. [DOI] [PubMed] [Google Scholar]
- 21.Nel, A., T. Xia, L. Mädler, and N. Li. 2006. Toxic potential of materials at the nanolevel. Science. 311:622–627. [DOI] [PubMed] [Google Scholar]
- 22.Jia, G., H. Wang, L. Yan, X. Wang, R. Pei, T. Yan, Y. Zhao, and X. Guo. 2005. Cytotoxicity of Carbon Nanomaterials: single-wall nanotube, multi-wall nanotube, and fullerene. Environ. Sci. Technol. 39:1378–1383. [DOI] [PubMed] [Google Scholar]
- 23.Hoet, P. H. M., I. Brüske-Hohlfeld, and O. V. Salata. 2004. Nanoparticles – known and unknown health risks. J. Nanobiotechnology. 2:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberdörster, G., A. Maynard, K. Donaldson, V. Gastranova, J. Fitzpatrick, K. Ausman, J. Carter, B. Karn, W. Kreyling, D. Lai, S. Olin, N. Monteiro-Rivere, D. Warheit, H. Yang, and A report from ILSI Research Foundation/Risk Science Institute Nanomaterial Toxicity Screening Working Group. 2005. Principles for characterizing the potential human health effects from exposure to nanomaterials: elements of a screening strategy. Particle Fibre Toxicol. 2:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, X., Y. Gui, R. M. Levenson, L. W. K. Chung, and S. Nie. 2004. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat. Biotechnol. 22:969–976. [DOI] [PubMed] [Google Scholar]
- 26.Bauer, L. A., N. S. Birenbaum, and G. J. Meyer. 2004. Biological applications of high aspect ratio nanoparticles. J. Mater. Chem. 14:517–526. [Google Scholar]
- 27.Mulvaney, S. P., M. D. Musick, C. D. Keating, and M. J. Natan. 2003. Glass-coated, analyte-tagged nanoparticles: a new tagging system based on detection with surface-enhanced Raman scattering. Langmuir. 19:4784–4790. [Google Scholar]
- 28.Knight, D. S., and W. B. White. 1989. Characterization of diamond films by Raman spectroscopy. J. Mater. Res. 4:385–393. [Google Scholar]
- 29.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, S.-J., M.-W. Kang, H.-C. Chang, K.-M. Chen, and Y.-C. Yu. 2005. Bright fluorescent nanodiamonds: no photobleaching and low cytotoxicity. J. Am. Chem. Soc. 127:17604–17605. [DOI] [PubMed] [Google Scholar]
- 31.Gaebel, T., I. Popa, A. Gruber, M. Domhan, F. Jelezko, and J. Wrachtrup. 2004. Stable single-photon source in near infrared. N. J. Phys. 6: article 98.
- 32.Chung, P.-H., E. Perevedentseva, and C.-L. Cheng. 2007. The particle size-dependent photoluminescence of nanodiamonds. Surf. Sci. doi:10.1016/j.susc.2007.04.150.
- 33.Schrand, A. M., H. Huang, C. Carlson, J. J. Schlager, E. Ohsawa, S. M. Hussain, and L. Dai. 2007. Are diamond nanoparticles cytotoxic? J. Phys. Chem. B. 111:2–7. [DOI] [PubMed] [Google Scholar]
- 34.Gruber, A., A. Dräbenstedt, C. Tiez, L. Fleury, J. Wrachtrup, and C. von Broczyskowski. 1997. Scanning confocal optical microscopy and magnetic resonance on single defect centers. Science. 276:2012–2014. [Google Scholar]
- 35.Kurtsiefer, C., S. Mayer, P. Zarda, and P. Weinfurter. 2000. Stable solid-state source of single photons. Phys. Rev. Lett. 85:290–293. [DOI] [PubMed] [Google Scholar]
- 36.Kühn, S., C. Hettich, C. Schmitt, J.-Ph. Poizat, and V. Sandoughdar. 2001. Diamond colour centers as a nanoscopic light source for scanning near field optical microscopy. J. Microsc. 202:2–6. [DOI] [PubMed] [Google Scholar]
- 37.Jelezko, F., C. Tietz, A. Gruber, I. Popa, A. Nizovtsev, S. Kilin, and J. Wrachtrup. 2001. Spectroscopy of single N-V centers in diamond. Single Mol. 2:255–260. [Google Scholar]
- 38.Iakoubovskii, K., and G. J. Adriaenssens. 2000. Luminescence excitation spectra in diamond. Phys. Rev. B. 61:10174–10182. [Google Scholar]
- 39.Lin, K.-W., C.-L. Cheng, and H.-C. Chang. 1998. Laser-induced intracluster reactions of oxygen-containing nanodiamonds. Chem. Mater. 10:1735–1737. [Google Scholar]
- 40.Jones, A. P., and L. d'Hendecourt. 2000. Interstellar nanodiamonds: the carriers of mid-infrared emission bands? Astron. Astrophys. 355:1191–1200. [Google Scholar]