Abstract
Drosophila N-cadherin (CadN) is an evolutionarily conserved, atypical classical cadherin, which has a large complex extracellular domain and a catenin-binding cytoplasmic domain. We have previously shown that CadN regulates target selection of R7 photoreceptor axons. To determine the functional domains of CadN, we conducted a structure-function analysis focusing on its in vitro adhesive activity and in vivo function in R7 growth cones. We found that the cytoplasmic domain of CadN is largely dispensable for the targeting of R7 growth cones, and it is not essential for mediating homophilic interaction in cultured cells. Instead, the cytoplasmic domain of CadN is required for maintaining proper growth cone morphology. Domain swapping with the extracellular domain of CadN2, a related but non-adhesive cadherin, revealed that the CadN extracellular domain is required for both adhesive activity and R7 targeting. Using a target-mosaic system, we generated CadN mutant clones in the optic lobe and examined the target-selection of genetically wild-type R7 growth cones to CadN mutant target neurons. We found that CadN, but neither LAR nor Liprin-α, is required in the medulla neurons for R7 growth cones to select sthe correct medulla layer. Together, these data suggest that CadN mediates homophilic adhesive interactions between R7 growth cones and medulla neurons to regulate layer-specific target selection.
Keywords: N-cadherin, neural target selection, layer-specific targeting, visual system development, adhesion molecule
Introduction
Cadherins (calcium-dependent cell adhesion receptors) provide intercellular adhesive interactions during a wide variety of developmental processes, including tissue morphogenesis and neurodevelopment (Gumbiner, 1996; Shapiro and Colman, 1999; Takeichi, 1991). The extracellular domains of cadherins contain various numbers of conserved cadherin repeats that mediate adhesion, while cytoplasmic domain sequences vary among different types of cadherins. In the classical type of cadherins, the intracellular domains contain two highly conserved regions: (1) the β-catenin-binding region, which links to the actin-cytoskeleton through α/β-catenins; and (2) the juxtamembrane region, which interacts with p120/δ-catenin (p120ctn) and other catenin family members (reviewed in Goodwin and Yap, 2004). Previous studies indicate that both cytoplasmic regions regulate cadherin adhesive activity via cytoskeleton regulators such as Rho-family GTPases (Braga et al., 1999). Recent studies have suggested that classical cadherins are not just passive adhesive molecules but rather adhesion-activated signaling receptors (Yap and Kovacs, 2003). In mammalian cell culture, E-cadherin has been shown to activate sequentially Rac GTPase, Src tyrosine kinase, and PI3-kinase, which, in turn, is responsible for efficient extension of cadherin-specific adhesive contact zones (Kovacs et al., 2002a; Roura et al., 1999). Inhibiting these signaling molecules using kinase inhibitors, or dominant negative constructs, disrupts the interplay between cadherin complexes and the actin-cytoskeleton. Even though cadherin-mediated signaling has been well demonstrated in cultured cells, investigation of its function in developmental contexts has been scarce, largely due to the pleiotropic functions of cadherins and the related signaling molecules (Owens et al. 2000; Noren et al. 2000). Using Drosophila border cell migration in oogenesis as a model, previous investigators have demonstrated that the β-catenin-binding site, and not the p120ctn-binding site, is critical for E-cadherin function in vivo ( Myster et al., 2003 ; Pacquelet et al., 2003 ). Yet, it is not known whether other classical cadherins function in the same fashion as E-cadherin. In this study, we used the single-cell mosaic technique to circumvent the pleiotropic functions of cadherins and investigated the requirement of cadherin-mediating signaling for making precise connections in the Drosophila visual system.
The Drosophila visual system has been widely used to study the molecular mechanism of neuronal connectivity due to the powerful genetic tools that are available in this system ( Ting and Lee, 2007 ; Clandinin and Zipursky, 2002 ; Tayler and Garrity, 2003 ). Fly compound eyes contain about 750 ommatidia, each of which has three types of photoreceptor neurons, R1-R6, R7, and R8 ( Meinertzhagen and Hanson, 1993 ). During development, each type of R-cells establishes layer-specific connections with its brain targets: R1-R6 connect to the first optic ganglion, the lamina, while R7 and R8 project axons through the lamina to terminate in two distinct layers in the second optic ganglion, the medulla ( Meinertzhagen and Hanson, 1993 ). A previous study reveals that Capricious, an adhesive molecule containing leucine-rich repeats, regulates R8 layer-specific targeting by promoting specific afferent-target interactions ( Shinza-Kameda et al., 2006 ). Two receptor tyrosine phosphatases, DPTP69D and LAR, as well as the adaptor protein Liprin-α, are required for R7s to establish layer-specific connections ( Hofmeyer et al., 2006 ; Choe et al., 2006 ; Maurel-Zaffran et al., 2001 ; Clandinin et al., 2001; Newsome et al., 2000 ). In a genetic screen based on visual behaviors, the neuronal cadherin (CadN) was identified for its requirement for R7 layer-specific targeting ( Lee et al., 2001 ). The Drosophila CadN belongs to a unique type of classical cadherin that is found in worms, insects, and vertebrates ( Tanabe et al., 2004 ), and are likely the most ancient form of classical cadherins ( Iwai et al., 2002 ; Iwai et al., 1997 ). Removing CadN in single R7 neurons resulted in R7 axons mistargeting to the R8-recipient layers, indicating that CadN functions cell-autonomously in R7s ( Lee et al., 2001 ). Recent developmental analysis further demonstrated that R7 neurons establish layer-specific connections in two separate developmental stages, and CadN is required for R7 growth cones to reach and remain in the R7-temporary layers in the first stage ( Ting et al., 2005 ). Although the requirement of CadN for R7 target selection is established, it remains unclear whether CadN provides adhesive interactions between R7 growth cones and their targets or mediates intracellular signaling processes that regulate the cytoskeleton in R7 growth cones. In this study, we address this issue by analyzing the ability of various CadN mutants to substitute for endogenous CadN activity.
Results
The CadN cyotoplasmic domain is required for proper growth-cone morphology and is largely dispensable for R7 target-selection
We have previously demonstrated that R7 axons project into their target layer in two distinct stages: during the early pupal stage (17% APF [after puparium formation]), R7 growth cones sequentially project into their temporary target layers where they remain until 50% APF when they regain motility to extend further into their destined layer (Figures 1A-A’’’’’) (Ting et al., 2005). Mosaic analyses using the GMR-Flp/MARCM system have revealed that CadN is required in R7s at both stages (Figures 1B-B’’’’’; Table 1; [Ting et al., 2005; Nern et al., 2005]). In addition, CadN mutant R7 growth cones often exhibit abnormal morphologies: they fail to expand in the target layer or expand prematurely before reaching the target layers (Figure 1B ’). The expression of full-length CadN protein in CadN mutant R7s, though, completely rescued these defects (Figures 1C -C’’’’’; Table 1). Based on this transgene-rescue system, we conducted a structure-function analysis to determine the necessity of the CadN cytoplasmic domain in R7 target selection.
Figure 1.
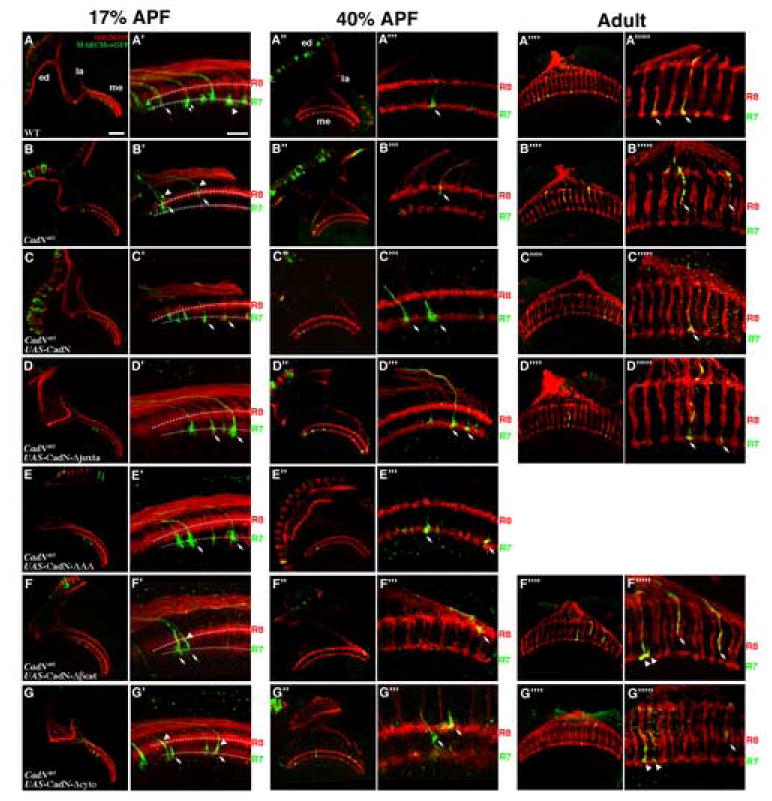
The intracellular domain of CadN is required for proper growth cone morphology, but is largely dispensable for R7 target selection.
R7 target selection was assessed in CadN mutant R7 clones with and without various transgene rescues. Single R7 cells, homozygous of a wild-type FRT40 chromosome (A-A””’) or CadN mutant chromosomes (B-B””’), were generated using GMR-Flp-mediated mitotic recombination, and labeled with mCD8-GFP or synb-GFP (green) using the MARCM system (A-B”’ or A””-B””’, respectively). All R-cell axons were visualized with MAb24B10 (red). Full-length CadN or CadN with various mutations were expressed in the CadN mutant R7 neurons to assess their ability to rescue R7 phenotypes (C-G””’). Layer-specific targeting and growth cone morphology were assessed at 17% APF (A-G’), 35% APF (A”-G’”) and at the adult stage (A””-G””’).
(A-A’) At 17% APF, newly differentiated photoreceptor neurons project axons into the anterior edge (to the left) of the medulla neuropil. The wild-type R7 growth cones follow the pioneering R8 axons to reach the R7-temporary layer (arrow), just beneath the R8 growth cones. The lamina growth cones (not shown) then project between R7 and R8 growth cones separating them into two layers (arrow head). (A”-A’”’’) At 35% APF and adult stage, the wild-type R7 and R8 growth cones form two separate layers (as indicated).
(B, B’) At 17% APF, approximately 21% CadN mutant R7 axons (arrows) fail to reach the R7-temporary layers; instead, they expand their growth cones incorrectly at the R8-temporary layer or between the R7- and R8-temporary layers. (B”, B’”) At 35% APF, about 55% of CadN mutant R7 axons terminate at incorrect layers. In addition, 63% and 19% of CadN mutant R7 growth cones fail to expand (arrowheads) at 17% and 35% APF, respectively. (B””, B””’) At the adult stage, essentially all CadN mutant R7 axons mistarget to the R8 layers or the layer between the R7 and R8 layers.
(C-C””’) Expressing full-length CadN in CadN mutant R7s fully rescues targeting and growth cone morphological defects (arrows).
(D-E”’) Expressing CadN with a deletion of the entire juxtamembrane domain (CadN-Δjuxta; D-D””’) or with a triple alanine mutation in the juxtamembrane domain (CadN-AAA; E-E’”) in CadN mutant R7s completely rescues the targeting phenotypes (arrows) albeit some (~20%) show minor morphological defects.
(F-G””’) CadN mutant R7s expressing CadN with a deletion of the β-catenin binding site (CadN-Δβcat; F-F””’), or a deletion of the entire cytoplasmic domain (CadN-Δcyto; G-G””’), target correctly at the R7-temporary layer at 17% APF (arrows; F, F’, G, G’). However, at 35% APF, some (12% and 16%) of the R7 growth cones retract to the distal layers, with ~20% of them exhibiting morphological defects (arrows; F”, F’”, G”, G’”). In the adults, 31% and 36% of these rescued R7 growth cones retract to the distal layers (arrows; F””, F””’, G””, G””’).
The presumptive R7- and R8-temporary layers are indicated by dotted lines in (A’-G’). (A’-G’, A”’-G”’ and A””’-G””’) High-magnification views of A-G, A”-G” and A””-G””, respectively. Scale bars: in A, 30 μm for A-G, A”-G” and A””-G””; in A’, 10 μm for A’-G’, A”’-G”’ and A””’-G””’.
Table 1.
Table summarizing the observed R7 targeting and morphological defects in wild-type, various mutant backgrounds, and transgene-rescues.
| Genotype | wild -type | CadN405 | CadN, CadN2 | CadN405 | CadN405 | CadN405 | CadN405 | CadN405 | CadN405 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Rescue Transgene | --- | --- | --- | UAS-CadN | UAS-CadN-AAA | UAS-CadNΔjuxta | UAS-CadNΔβ-cat | UAS-CadNΔcyto | UAS-CadN2-CadN chimera | |
| 17%APF | Mistargeting | 0%(n=40) | 21%(n=43) | 41%(n=90) | 0%(n=75) | 0%(n=27) | 0%(n=59) | 0%(n=61) | 0%(n=44) | 55%(n=118) |
| Growth ConeDefects | 3%(n=40) | 63%(n=43) | 94%(n=90) | 6%(n=75) | 21%(n=27) | 20%(n=59) | 33%(n=61) | 40%(n=44) | 85%(n=118) | |
| 35%APF | Mistargeting | 0%(n=26) | 55%(n=26) | 53%(n=114) | 0%(n=38) | 0%(n=52) | 0%(n=60) | 12%(n=74) | 16%(n=50) | 64%(n=69) |
| Growth Cone Defects | 0%(n=26) | 19%(n=26) | 89%(n=114) | 8%(n=38) | 7%(n=52) | 3%(n=60) | 23%(n=74) | 22%(n=50) | 51%(n=69) | |
| Adult | Mistargeting | 0%(n=66) | 100%(n=59) | 100%(n=37) | 0%(n=64) | N/A | 0%(n=63) | 31%(n=142) | 36%(n=109) | N/A |
The CadN cytoplasmic domain contains two regions: (1) the juxtamembrane region, which, it has been proposed, interacts with p120 or other catenins; and (2) the β-catenin binding region (Iwai et al., 1997). We found that the juxtamembrane region is not required for CadN function in R7 target selection. The expression of the CadN mutants with triple-alanine mutations in the presumptive p120 binding site (CadN-AAA), or a deletion of the entire juxtamembrane region (CadN-Δjuxta) completely rescues the CadN mutant R7 targeting phenotype (Figures 1 D-E”’ and Table 1). Expressing CadN-AAA or CadN-Δjuxta partially rectified the morphological defects of the CadN mutant growth cones at 17% APF (21% and 20% defective, respectively), and fully rescued these defects at 35% APF (7% and 3% defective, respectively). Similarly, the expression of the CadN mutants with a deletion of either the β-catenin binding region (CadN-Δβcat) or the entire cytoplasmic domain (CadN-Δcyto) completely rescues R7 targeting defects at 17% APF. On the other hand, with CadN-Δβcat and CadN-Δcyto, 12% and 16% respectively of the R7 axons fail to remain in the R7-temporary layer at 35% APF, and 31% and 36% respectively of R7s mistarget to the R8 layer in the adult (Figures 1 F-G””’ and Table 1). In addition, a substantial percentage of CadN mutant growth cones expressing CadN-Δβcat (33% and 23%) or CadN-Δcyto (44% and 22%) still exhibited collapsed morphology at 17% and 35% APF. Together, the rescue results suggest that the cytoplasmic domain is dispensable for initial R7 target selection, but the β-catenin binding region does contribute to the ability of R7 growth cones to remain and expand properly in the target layer.
The CadN2 gene encodes for multiple alternative transcripts
The Drosophila genome contains a CadN–homologous gene, CadN2, which is located next to the CadN gene and most likely originated from a partial duplication of the CadN gene (Figure 2A) (Prakash et al., 2005). The CadN2 gene is homologous to the CadN gene (amino-acid sequence identity 72.5%), but the predicted protein sequence contains an extracellular domain smaller than that of the CadN protein (Figure 2B). We further characterized CadN2 function and conducted a comparative structure-function analysis focusing on the extracellular domain (see below).
Figure 2. Exon organization of the Drosophila CadN2 gene.
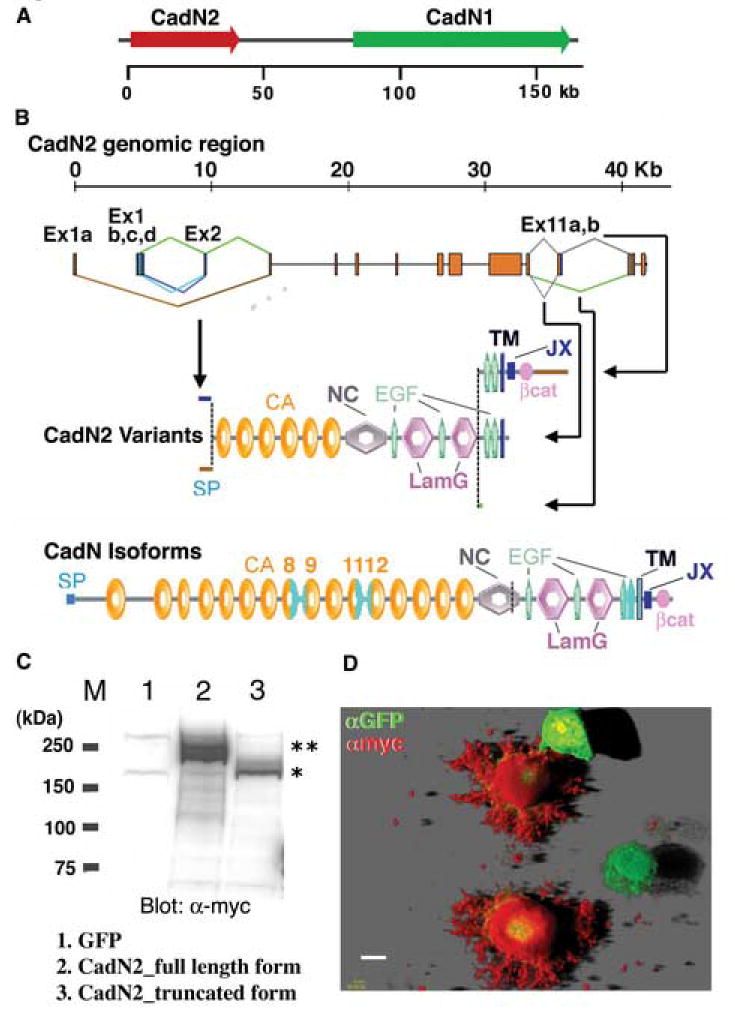
(A) The Drosophila genome contains a CadN–homologous gene, CadN2, which is located next to the CadN gene in a head-to-tail orientation. (B) The CadN2 gene contains 14 exons that span approximately 40 Kb of the genomic DNA. Complex alternative splicing occurs in exons 1, 2, and 11. As a result of the alternative use of exons 1a or 1bcd/2, mature CadN2 mRNAs contain variable 5’ sequences as well as different 5’ splice sites of exon 1bcd. Exons 1a and 2 encode two different signal peptide sequences. Alternative use of the exon 11a,b and its 5’ splice site results in three types of transcripts: (i) one that contains the exon 11a and encodes a receptor form; (ii) another that misses the 5’ splice site of the exon 11a to terminates shortly afterward and encodes a truncated form; and (iii) the last that omits the exon 11 and encodes a secreted form. Constant exons are shown as red boxes and alternative exons as green, blue, or brown boxes. SP: signal peptide; CA: cadherin repeat; NC: noncordate domain; EGF: EGF-like calcium-binding repeat; LamG: Laminin-G-like domain; TM: transmembrane domain; JX: juxtamembrane region; βcat: β-catenin binding region. The domain structure of CadN isoforms is shown for comparison. Variable regions in the cadherin and transmembrane domains of CadN are colored in light blue. (C) Western blot analysis of CadN2 protein expressed in S2 cells. Extracts of S2 cells expressing GFP (lane 1), the myc-tagged CadN2 receptor forms (lanes 2), and the myc-tagged CadN2 truncated forms (lane 3) were analyzed by Western blot and probed with anti-myc antibody. A myc tag was used to mark the C-termini of CadN2 proteins. Doublets of ~220 kDa (lane 2) and ~200 kDa (lane 3) detected by anti-myc corresponds to the receptor (predicted molecular weight ~200 kDa) and truncated forms of CadN2 protein (predicted molecular weight ~180 kDa) with variable glycosylation, respectively. M: molecular mass markers (in kDa). (D) Confocal image of the S2-adhesion cells expressing the myc-tagged CadN2 receptor form and a GFP marker. CadN2 protein and GFP were visualized using anti-myc (red) and anti-GFP antibodies (green), respectively. Note that CadN2 protein was detected on the cell surface and the cell processes. In the figure, two S2 cells expressing only GFP were not labeled by anti-myc antibody. Scale bar: 10 μm.
We cloned multiple CadN2 cDNAs, which were subsequently mapped onto the genomic sequence (see Materials and methods for details). These analyses reveal that the CadN2 gene undergoes alternative splicing and can generate potentially 12 different transcripts that are predicted to translate into six different protein variants (Figure 2B). All the CadN2 variants predicted from the cDNA sequences contain six cadherin repeats, four EGF-CA domains, and two Lam-G domains in their extracellular domain as well as three different carboxyl-termini, resulting in (1) a full receptor form; (2) a cytoplasmic-domain-truncated form; and (3) a secreted form that lacks the transmembrane domain. Although all the alternative exons were found in the RNA isolated from developing eye discs, we recovered most frequently the CadN2 cDNA containing exons 1d and 11a in the 5’ and 3’ variable regions. This variant, then, was used for subsequent analysis unless otherwise stated.
CadN2 plays an accessory role in R7 target selection
Because CadN2 is expressed in the eye discs during the R7 target-selection period, we set out to determine whether CadN2 plays a role in R7 target selection. First, we examined the homozygous CadN2 null mutants (Δ7) and the CadN2/CadN-CadN2(Δ4) (a deficiency uncovered in both CadN and CadN2 genes) hetero-allelic mutants ( Prakash et al., 2005 ). We found these mutant animals to be viable and without detectable R7 targeting defects, indicating that CadN2 is not essential for R7 target selection (data not shown). We next examined target selection in single CadN CadN2 double mutant R7s using the GMR-Flp/MARCM system. We found that CadN CadN2 double mutant R7 axons exhibit mistargeting phenotypes and growth cone morphological defects similar to those in CadN mutants (Figures 3A -B’). However, a significantly higher percentage of CadN CadN2 double mutant R7s exhibits both targeting and growth cone morphological defects than do CadN mutants at 17% and 35% APF (Table 1). In addition, CadN CadN2 double mutant R7s exhibit phenotypes that have not been observed in CadN mutants: approximately 20% of CadN CadN2 double mutant R7s overshoot the R7-temporary layer and project into the presumptive inner medulla, and some 26% of the growth cones expand abnormally in both the R7 and R8 layers ( supplementary Figures 1A-C). The pleiotropy phenotypes of the CadN CadN2 double mutants might reflect the complexity of CadN2 variants expressed in the R7s. These phenotypes, however, can be completely rescued by expressing the wild-type CadN or, to a lesser extend, by expressing CadN-Δcyto (38% mistargeting) (Figures 3C-C’). In summary, CadN2 is partially redundant to CadN in regulating R7 target selection.
Figure 3.
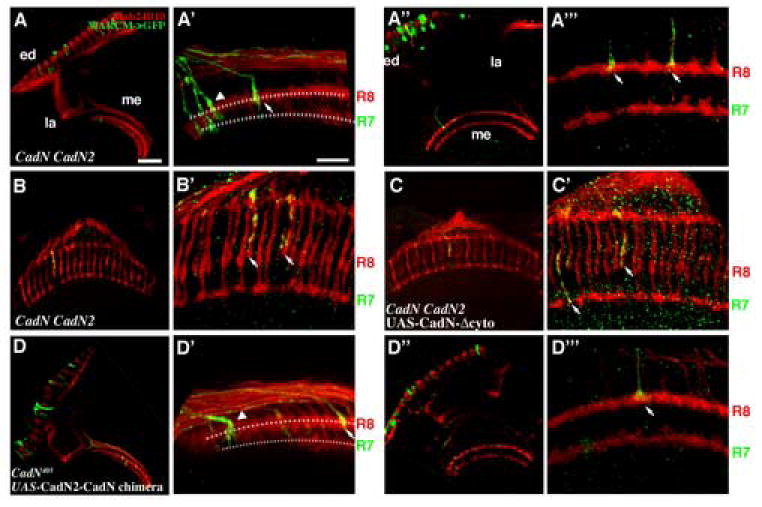
CadN2 is partly redundant to CadN in R7 target selection. Mosaic CadN CadN2 mutant R7 cells were generated using GMR-Flp and labeled with mCD8-GFP or synb-GFP (green) using the MARCM system. All R-cell axons were visualized with MAb24B10 (red). (A, A’) At 17% APF, approximately 41% of the CadN CadN2 double mutant R7 axons (arrows) terminated incorrectly at the R8-temporary layer or between the R7- and R8-temporary layers. (A’’, A’”) At 35% APF, about 53% of the CadN CadN2 double mutant growth cones terminated at incorrect layers and exhibited collapsed morphology (arrowheads), as seen in CadN mutant R7 growth cones. (B, B’) In the adult, essentially all of the CadN CadN2 double mutant axons (arrows) terminated incorrectly at the R8 layer. In addition, approximately 20% of the CadN CadN2 double mutant R7 axons exhibited novel phenotypes that were not seen in CadN mutants ( supplementary Figures 1 B and C). (C, C’) Expressing CadN-Δcyto in CadN CadN2 double mutant R7 partially rescued the mistargeting phenotypes (approximately 38% mistargeting in adults). (D-D’”) Expressing CadN2-CadN chimera protein in CadN mutant R7s exacerbated R7 targeting and growth cone morphological defects. Approximately twice as many R7 axons mistargeted and exhibited collapsed growth cones as seen in CadN mutants (arrowheads) at the 17 hr (D, D’) and 35 hr AFP (D”, D”’). The presumptive R7- and R8-temporary layers are indicated by dotted lines in (A’, D’). (A’, A’’’, B’, C’, D’, D’’’) High-magnification views of A, A’’, B, C, D and D”, respectively. Scale bars: in A, 30 μm for B, D, A’’, C and D’’; in A’, 10 μm for B’, D’, A’’’, C’ and D’’’.
The CadN cytoplasmic domain is not required for in vitro adhesive activity
Previous studies suggest that the cytoplasmic domain of classical cadherins regulates cell adhesive activity via interactions with catenins and other cellular signaling molecules (Braga et al., 1999). Thus, we used an S2 cell-aggregation assay (Oda et al., 1994; Ting et al., 2005) to examine whether removing different regions of CadN cytoplasmic domain affects its adhesive activity. In these experiments, S2 cells expressing various CadN cytoplasmic-deletion mutants were assayed for their ability to induce cell aggregates. We found that all the CadN mutants with various deletions of the cytoplasmic domains induce cell aggregations (Figures 4B-E). We quantified the sizes of the cell aggregates induced by CadN lacking the entire cytoplasmic domain, and found that they are essentially indistinguishable from those induced by full-length CadN (Figure 4H; see Materials and methods for details). These data indicate, as previously shown for Drosophila E-cadherin ( Pacquelet et al., 2003 ), that the CadN cytoplasmic domain is not required for homophilic adhesive activity in vitro.
Figure 4.
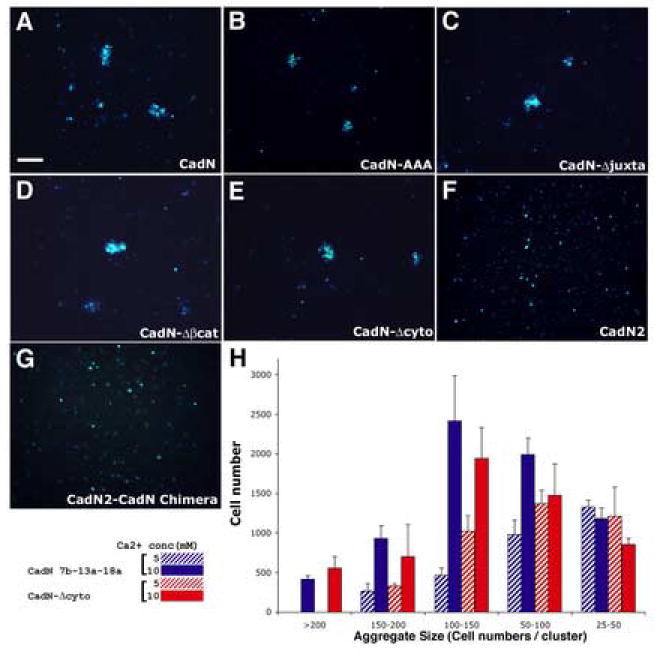
CadN extracellular domain is required and sufficient for mediating homophilic interactions in vitro. S2-suspension cells expressing full-length CadN (A), CadN-AAA (B), CadN-Δjuxta (C), CadN-Δβcat (D), CadN-Δcyto (E), CadN2 (F), or CadN2-CadN chimera (G) were assessed for their ability to induce cell aggregation in the presence of 10 mM calcium. A GFP marker (blue) was used to label the transfected S2 cells. The S2 cells expressing full-length CadN (A) or cytoplasmic mutants (B-E) form cell clusters. In contrast, the S2 cells expressing CadN2 or CadN2-CadN chimera are dispersed (F, G). (H) A bar chart of the size of the cell aggregates formed by S2 cells expressing full-length CadN (blue bars) and CadN-Δcyto (red bars) in the presence of 5 (stripe bars) or 10 (solid bars) mM of calcium. Cell aggregates are divided into five categories according to the number of cells in each aggregate. X-axis: cell-aggregate size (25-50, 50-100, 100-150, 150-200, and greater than 200 cells per cell cluster). Y-axis: the total number of cells that form a given range of cell cluster size. Bars: means of three independent results. Error bars: standard error. Scale bar in A, 100 μm.
The CadN’s extracellular domain cannot be substituted by that of CadN2 for homophilic interactions in vitro and R7 target selection in vivo
We next examined whether the CadN2 protein mediates homophilic interactions using the S2 cell-aggregation assay. We found that neither the receptor form nor the cytoplasmic-domain truncated form is able to induce cell aggregates when expressed in S2 cells (Figure 4 F and data not shown). To confirm the expression of CadN2 protein in S2 cells, we engineered a myc-tag at the carboxyl-terminus of CadN2 and expressed the tagged CadN2 proteins in S2 cells. Like their untagged counterparts, myc-tagged CadN2 proteins also do not induce cell aggregates (data not shown). Nonetheless, Western blot and immunohistochemistry analyses confirm that both the receptor and cytoplasmic-domain truncated forms are readily expressed and present at the cell surface (Figures 2 C and D). In conclusion, CadN2 does not mediate homophilic interaction in vitro.
To determine if the lack of adhesive activity of CadN2 is dictated by its extracellular domain, we generated a CadN2-CadN chimera construct, which encodes the CadN2 extracellular domain and the CadN transmembrane and cytoplasmic domains. CadN2-CadN chimera when expressed in S2 cells failed to induce cell aggregation, indicating that the CadN2 extracellular domain is not capable of mediating homophilic interactions. We next examined whether CadN2-CadN chimera can rescue CadN mutant R7 phenotypes. Expressing CadN2-CadN chimera in CadN mutant R7s results in more severe targeting and growth cone morphological defects than those observed in CadN mutant R7s (Figures 3D-D’’’; Table 1). The exacerbated phenotype indicates that CadN2-CadN chimera not only fails to be a substitute for CadN function in R7 growth cones, but also actively disrupts R7 targeting. The seemly dominant-negative activity of CadN2-CadN is only observable in the CadN mutant background: the expression of CadN2-CadN in the wild-type background does not disrupt R7 targeting significantly ( supplementary Figures 2B ,B’). In summary, the CadN2 extracellular domain cannot be substituted for the CadN extracellular domain for mediating cell adhesion in vitro and for R7 target selection in vivo.
CadN, but not LAR or Liprin-α, is required in the target neurons for R7-target selection
Previous studies indicated that R7 target selection is dictated by the interactions between R7 afferents and medulla targets. Since the CadN extracellular domain, which mediates homophilic interaction, is required for R7 target selection, we set out to determine whether CadN mediates adhesive interactions between R7 axons and medulla neurons. We generated CadN mutant medulla clones using a recently developed mosaic system, the ELF system (ey-Gal80,lama-Gal4, UAS-Flp system; [ Chotard et al., 2005 ]). In this system, flipase was expressed specifically in lamina and medulla precursor cells to induce mitotic recombination in these cells, and the mosaic clones were marked by the lost of GFP signal. Using this system, we generated mosaic animals that had approximately 30-40% of their medulla neurons rendered homozygous CadN mutant while their retina and the rest of the animals were heterozygous and presumably wild-type. We first examined their R-cell connection patterns in the adult stage. We found that these CadN mosaic animals, unlike their wild-type controls (Figures 5A-A’’’), had severe retinotopic mapping defects: the R-cell axons frequently crossed over one another and some of them projected further into the distal medulla neuropil (Figures 5B -B’’’). Using R7 and R8 specific markers, we found that layer-specific targeting of R7 and R8 axons was disrupted in these animals: some R7 axons terminated incorrectly in the R8 layer, and some R8 axons projected deeper into the R7 layer or the layers below. In addition, the R7 and R8 termini exhibited abnormal morphologies. Removing both CadN and CadN2 in the target neurons resulted in seemingly severer phenotypes (Figures 5C -C’’’). However, in the presence of severe retinotopic mapping defects, we were not able to quantify the layer-specific targeting defects in great precision.
Figure 5.
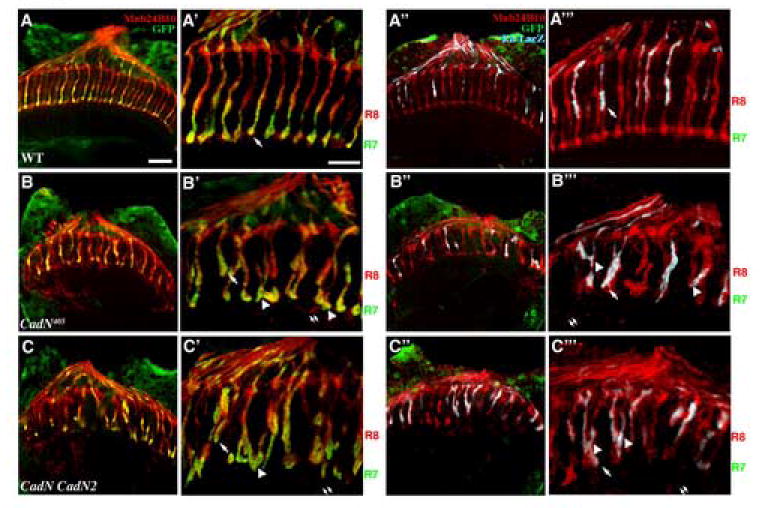
CadN is required in the optic lobe for R7 and R8 layer-specific targeting. Approximately 40% of the medulla neurons were rendered homozygous of a wild-type FRT40 (A-A”’) or CadN mutant (B-B”’) or CadN CadN2 double mutant (C-C”’) chromosomes using the ELF system (see Materials and methods for details). The mosaic tissues were marked by the loss of GFP signal. The layer-specific targeting of R7 and R8 axons were assessed in adult animals, using PanR7-Gal4, UAS-mCD8-GFP (green) (AC’) and Rh5-Gal4, UAS-lacZ (blue)(A”-C”’), respectively. MAb24B10 (red) was used to label all R-cell axons. (A-A”’) In the wild-type control, both R7 and R8 axons targeted to correct layers (arrow). (B-B”’) CadN medulla-mosaic animals exhibited severe retinotopic mapping defects: the R-cell axons frequently crossed over one another with some of them projecting further into the proximal medulla neuropil. Some R7 axons terminated incorrectly in the R8 layer (arrow, B’); many R8 axons projected deeper into the R7 layer (arrows, B”’) or the layers below (double arrows, B”’). In addition, the R7 and R8 axonal termini exhibited abnormal morphologies with some R-cell axons extending processes into proximal layers (double arrows). (C-C”’) Removing both CadN and CadN2 in the medulla neurons resulted in seemingly severer phenotypes. A’-C’ and A”’-C”’ are high magnification views of A-C and A”-C”, respectively. Scale bars: in A, 30 μm for B-C, and A”-C”; in A’, 10 μm for B’-C’ and A”’-C”’.
We have previously demonstrated that the receptor tyrosine, LAR, is required in R7s for target selection. Recently, we and others found that removing Liprin-α, a cytosolic partner of LAR, in the R-cells resulted in LAR-like R7-mistargeting phenotypes (Gao and Lee, unpublished observation; [Hofmeyer et al., 2006; Choe et al., 2006]). We set out to examine whether LAR or Liprin-α is required in the target neurons for R7 target selection. Using the ELF system, we generated LAR or Liprin-α target mosaic animals and found that R7 and R8 connections are essentially indistinguishable from the wild-type controls (supplementary Figures 3B -C). This indicates that, in contrast to CadN, LAR and Liprin-α are dispensable in the medulla and lamina neurons for R7 target selection and topographic mapping.
CadN is required in the target neurons for the initial targeting of R7 axons
To determine when the connectivity defects occur in CadN target-mosaic animals, we examined R7 target selection during development. In wild-types, at the 17% APF, newly differentiated R7 project axons into the medulla neuropil to terminate at the layer just below the R8 growth cones (the R7-temporary layer); after 2-4 hrs, the axons of newly differentiated lamina neurons follow R7 axons and project between R7 and R8 growth cones separating them into two layers (Figures 6A, A’; [Ting et al., 2005]). In the CadN or CadN CadN2 mutant target-mosaic animals, genetically wild-type R7 axons projected into the medulla neuropil made by mutant medulla neurons. However, many of them (33.7 %) failed to project past R8 growth cones (Figures 6B , B’, C, C’), indicating that CadN is required in the medulla for R7 target selection at the first stage. The retinotopic map and medulla neuropil organization (as assessed by HRP and Capricious staining) appears to be unaffected in the CadN target-mosaic animals, suggesting that CadN is not required in the medulla neurons for early developmental processes ( supplementary Figure 4). At 35% APF, more growth cones from lamina neurons and processes from medulla neurons extended into the region between R7 and R8 growth cones to further separate these two growth cone layers (Figures 6A”, A’”). In the CadN or CadN CadN2 target-mosaic animals, R7 and R8 axons exhibited severe topographic mapping and layer-specific targeting defects (Figures 6B ”, B’”, C”, C’”). At 35% APF, these R7 and R8 axons frequently crossed over one another and target to incorrect layers, as seen in the adult stage. The exacerbation of phenotypes during the passive displacement period suggests that CadN mutant lamina and medulla neurons actively disrupt R7 and R8 connections.
Figure 6.
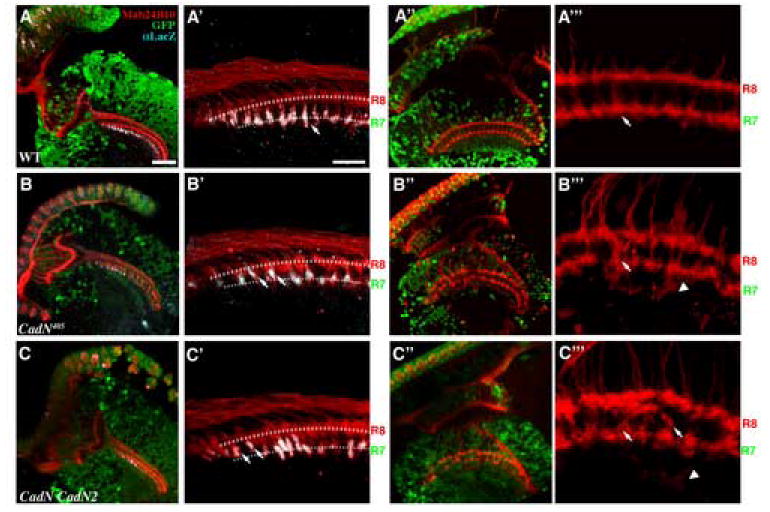
CadN is required in medulla neurons for R7 targeting in the first layer-selection stage. Optic lobes were rendered wild-type (A-A”’), CadN (B-B”’), or CadN CadN2 double mutant (C-C”’) using the ELF system; the mosaic neurons were marked by the loss of GFP signal. (A-C’) The initial projections of R7 axons into the R7-temporary layer were assessed at 17% APF. R7 axons were labeled using PM181-LacZ (grey) and R-cell axons were visualized with MAb24B10 (red). (B, B’ and C, C’) In the CadN or CadN CadN2 target-mosaic animals, genetically wild-type R7 axons projected into the medulla neuropil, composed of processes of mutant medulla neurons. Unlike in wild-type (A-A’), many of the CadN mutant R7s axons (33.7 %) fail to project past R8 growth cones (arrows). (B”, B”’ and C”, C”’) At 37% APF, R7 and R8 axons exhibit severe topographic mapping and layer-specific targeting defects (arrows) in the CadN or CadN CadN2 mutant target-mosaic animals. Moreover, some R-cell axons extend processes to deeper layers (arrowheads). A’-C’ and A”’-C”’ are high magnification view of A-C and A”-C”, respectively. The presumptive R7- and R8-temporary layers are indicated by dotted lines in (A’-C’). Scale bars: in A, 30 μm for B-C and A”-C”; in A’, 10 μm for B’-C’ and A”’-C”’.
Discussion
CadN mediates adhesive interactions between R7 growth cones and the processes of medulla neurons
We have previously demonstrated that CadN functions in the first target-selection stage for R7 axons to reach and remain in the R7-temporary layer (Ting et al., 2005). In this study, we determine that CadN’s function in this process depends on its extracellular and not its cytoplasmic domain: CadN cytoplasmic deletion mutants but not CadN2-CadN chimera largely rescue R7 targeting defects. The CadN extracellular domain provides its adhesive activity: all the CadN cytoplasmic-domain deletion mutants, but not CadN2 or CadN2-CadN chimera, mediate homophilic interaction in vitro. Taken together, the data suggests that it is the adhesive and not the signaling activity of CadN that plays the key role in the initial targeting of R7s. The importance of adhesive interaction for initial axonal targeting is underscored by the observation that the removal of CadN in medulla neurons disrupted the initial targeting of R7 axons. In vivo studies in grasshoppers and flies have demonstrated that growth cones extend highly dynamic filapodia which, when encountering appropriate substrates or targets, become stabilized and dilated and could eventually “pull” the growth cones toward the targets (Murray et al., 1998; O′Connor and Bentley, 1993; Ritzenthaler et al., 2000). Similarly, we have observed R7 growth cones extend CadN-expressing filapodia into the R7 temporary layer during the initial targeting, ([Ting et al., 2005], Yonekura and Lee, unpublished observation). We envisioned that CadN provides the initial adhesive interaction between R7 filapodia and temporary target and stabilizes the contacts (Figure 7).
Figure 7.
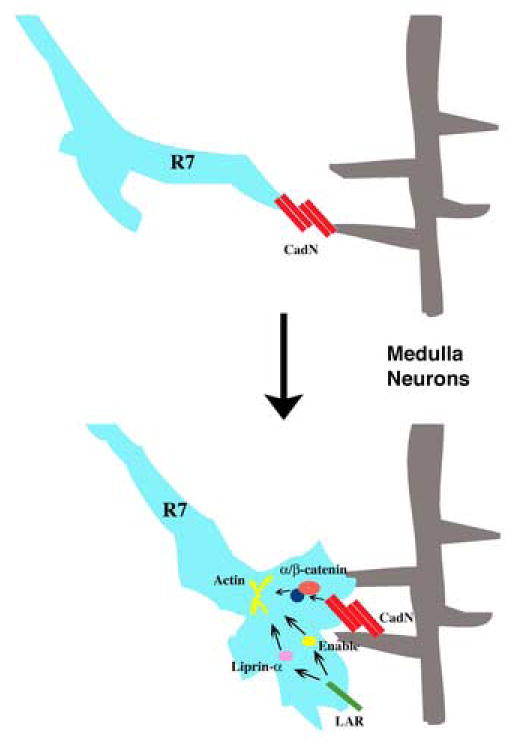
A schematic model for CadN’s function in R7 target selection. CadN mediates adhesive interactions between R7 growth cones and neurites of the medulla neurons during the first target selection stage. The CadN cytoplasmic domain interacts with α/β-catenins to regulate growth cone morphology. LAR and Liprin-α function in R7 growth cones, presumably to regulate actin-cytoskeleton for R7 target selection.
We have previously shown that removing CadN in a small number of medulla neurons does not affect R7 target selection (Ting et al., 2005). In this study, the ELF mosaic system was used to generate large and widespread clones of CadN mutant medulla neurons; a procedure which resulted in R7 target selection defects. The difference between these two observations likely reflects that multiple medulla neurons can serve as redundant targets for R7 growth cones. Similar target redundancy was recently reported in lamina neurons for R1-6 target selection ( Prakash et al., 2005 ). Based on Golgi staining, the medulla is estimated to contain some fifty neuron subtypes. The identities of the R7 temporary targets in the medulla have yet to be determined. By screening a large collection of Gal4 enhancer trap lines, we identified a group of apterous-expressing medulla neurons that extend their processes in the R7-temporary layer during the first stage of R7 target selection ( supplementary Figures 5A-A”’). This group of neurons expresses CadN, therefore, it could potentially serve as R7 target neurons during development (supplementary Figures 5A-B’). They do not appear, however, to synapse with R7 neurons in adults, suggesting that they only function as temporary targets (S.-Y. Gao and C.-H. Lee, unpublished observation). In the vertebrate hippocampus, entorhinal axons form temporary synapses with Caja-Retzius cells and later move to synapse with their destined targets, the pyramidal neurons. We have previously reported that R7 axons remain in the temporary target layer for some twenty hours during which the medulla undergoes significant development. We speculate that the first targeting step serves to coordinate afferent innervation with target development: during the second stage, R7 axons regain motility and target to the destined synaptic partners only after these target neurons become fully developed.
In this study, we demonstrated that CadN2 functions partially redundantly to CadN in regulating R7 target selection at the first stage. Bases on its lack of adhesive activity in culture cells, CadN2 likely has functions that are distinct from that of CadN. Interestingly, expressing the CadN2-CadN chimera in CadN mutant R7s exacerbates CadN phenotypes, suggesting that CadN2-CadN activates cytoplasmic signaing in an abnormal context. Previous studies in vertebrate systems have revealed that cadherins regulate FGF-mediate signaling (Lom et al., 1998) and integrin-mediated adhesion (Lilien J., 1999). We speculate that CadN2 couples with yet-to-be identified adhesive molecules or signaling receptors that function redundantly with CadN.
The cytoplasmic domain of CadN is required for regulating growth cone morphology
Previous studies have demonstrated that classical cadherins function not only as adhesive molecules but also as signaling receptors via their catenin-binding cytoplasmic domains (Yap and Kovacs, 2003). Our structure-function analysis revealed that the CadN-mediated signaling is largely dispensable for initial target selection; instead, it is required for regulating growth-cone morphology. In wild-types, R7 growth cones change from motile spare-like structures to fully-expanded growth cones after they reach the target region, and this morphological conversion is partially dependent on the CadN cytoplasmic domain. The CadN cytoplasmic domain has been shown to interact with β-catenin and likely p120/δ-catenin (or other signaling molecules), and these interactions could potentially regulate the actin-cytoskeleton (Anastasiadis and Reynolds, 2001; Kovacs et al., 2002b; Noren et al., 2000). Previous studies reveal that growth cones undergo drastic morphological changes when they encounter different adhesion molecules in the substrate (Payne et al., 1992; Burden-Gulley et al., 1995). We speculate that the morphological changes at the first target selection stage – from a spear-like structure to a fully expanded shape – reflects that R7 growth cones migrate to different substrates, i.e., from R8 axons to the dendrites of the medulla target neurons. This migration likely sets up R7 growth cones for the second target selection stage when they must proceed, in the absence of guidance of R8, to their final target layer.
While both β-catenin and p120/δ-catenin binding sites regulate growth morphologies, the β-catenin binding site appears to have an additional function. The CadN mutant R7 growth cones expressing CadN-Δβcat reached the correct targets initially, but some (12%) axons retracted to more superficial layers at 35% APF. During this period, lamina axons projected between the R7 and R8 growth cones to form new intervening layers while R7 and R8 growth cones had to hold on to their temporary targets (Ting et al., 2005). Thus, CadN-Δβcat might not provide proper adhesive force for R7 growth cones to remain in the R7-temporary layer. Alternatively, the β–catenin binding region of CadN might, in response to a target-derived cue, regulate actin-cytoskeletal structure and/or morphology of R7 growth cones for remaining in the R7-temporary layer.
Interestingly, CadN mutant R7s rescued by CadN-Δβcat or CadN-Δcyto exhibit similar, but weaker phenotypes to those of LAR or Liprin-α mutants. Previous studies in vertebrate systems demonstrated physical association between vertebrate LAR-family receptor tyrosine phosphatases and N-cadherin-β-catenin complexes ( Kypta et al., 1996 ; Brady-Kalnay et al., 1998 ). Recently, Drosophila CadN and LAR have been found to interact genetically ( Clandinin T.R. et al. , personal communication), suggesting a crosstalk between CadN- and LAR-mediated pathways. However, our attempt to establish an epistatic relationship between CadN and LAR was not successful: overexpression CadN in LAR mutant background failed to rescue LAR phenotypes and vice versa (Yonekura and Lee, unpublished observation). Further dissection of CadN-LAR physical interaction and Liprin-α function would be needed to resolve the complex crosstalk between these two pathways. Nonetheless, our finding that expressing a CadN mutant lacking the entire cytoplasmic region can largely rescue CadN mutant phenotype underscores the importance of CadN adhesive activity in regulating neural target selection.
Materials and methods
Genetics
CadN405, CadN2(Δ7), CadN CadN2(Δ4) , and LAR451 mutants have been described previously ( Lee et al., 2001 ; Prakash et al., 2005 ). The Liprin-α1 mutant was isolated in a previous described genetic screen ( Lee et al., 2001 ) and characterized by Choe et al. ( Choe et al., 2006 ). Genetic mosaic R7 neurons were generated using the GMR-Flp/MARCM system and targeting was analyzed at 17%, 35% APF and at the adult stage as described ( Ting et al., 2005 ). Fly stocks used for these experiments are as follows: (1) GMR-Flp; FRT40; (2) GMR-Flp; CadN405FRT40/CyO, ubiP-GFP; (3) GMR-Flp; CadN CadN2 (Δ4) FRT40/CyO, ubiP-GFP; (4) GMR-Flp, UAS-CadN7b-13a-18a; CadN405FRT40/CyO ubiP-GFP; (5) GMR-Flp; CadN405FRT40/CyO, ubiP-GFP; UAS-CadN7b-13a-18a-AAA/TM6b; (6) GMR-Flp; CadN405FRT40/CyO, ubiP-GFP; UAS-CadN7b-13a-18a-Δjuxta; (7) GMR-Flp; CadN405FRT40/CyO, ubiP-GFP; UAS-CadN7b-13a-18a-Δβcat; (8) GMR-Flp; CadN405FRT40/CyO, ubiP-GFP; UAS-CadN7b-13a-18a-Δcyto; (9) GMR-Flp; CadN405FRT40/CyO, ubiP-GFP; UAS-CadN2-CadN7b-13a-18a; (10) Elav-Gal4c155, UAS-mCD8-GFP; tubP-Gal80 FRT40
Mutant medulla neurons were generated using a previously published mosaic system, the ELF system (Chotard et al., 2005). In this system, flipase is expressed specifically in lamina and medulla precursor cells to induce mitotic recombination in these cells and the mosaic clones were marked by the loss of GFP signal. Using this system, we generated mosaic animals that had approximately 30-50% of their medulla neurons rendered homozygous CadN mutant or CadN CadN2 double mutant while their retina and the rest of the animal remained heterozygous and presumably wild-type. Fly stocks used for these experiments are as follows: (1) ey-Gal80; ubiP-GFP CycE FRT40/GlaBc; Lama-Gal4 UAS-Flp/TM6b; (2) w; CadN405FRT40/CyO, ubiP-GFP; PanR7-Gal4 UAS-GFP/TM6b; (3) w; CadN CadN2 (Δ4) FRT40/CyO, ubiP-GFP; PanR7-Gal4 UAS-GFP/TM6b; (4) w; CadN405FRT40/CyO, ubiP-GFP; Rh5-Gal4 UAS-LacZ/TM6b; (5) w; CadN CadN2 (Δ4) FRT40/CyO, ubiP-GFP; Rh5-Gal4 UAS-LacZ/TM6b; (6) w; CadN405FRT40/CyO, ubiP-GFP; PM181-LacZ/TM6b; (7) w; CadN CadN2 (Δ4) FRT40/CyO, ubiP-GFP; PM181-LacZ/TM6b; (8) w; LAR451FRT40/CyO, ubiP-GFP; (9) w; Liprin-α1FRT40/CyO, ubiP-GFP
Molecular Cloning of the CadN2 gene
cDNA library screening, RT-PCR, and 5’ and 3’ RACE were used to clone the CadN2 cDNAs. We uncovered four different 5’ ends of CadN2 cDNAs, which appear to be the results of the alternative use of two transcription starting sites, three 5’splice sites of exon 1bcd, and exon 2 (Figure 2). The exons 1a and 2 encode for two different amino-terminal sequences that might serve as signal peptides. The transcripts containing exons 1d and 2 are the most abundant forms in developing eye discs and were used for subsequent analyses. In addition, we found three different 3’ ends generated by the alternative use of exon 11ab and its 5’ splice site. These translate into three different carboxyl-terminal sequences: one predicted to encode a receptor form, a cytoplasmic truncated form, and a secreted form. Apart from the variable regions, all CadN2 variants are predicted to contain six cadherin repeats, two EGF-CA repeats, and two Lamin-like globulin domains in their extracellular domains. CadN2 cDNA sequences are being submitted to Flybase. Bioinformatic analysis revealed CadN2 homologous genes in Drosophila pseudoobscura, another member of the Drosophila genus, and the malaria mosquito, Anopheles gambiae, both of which diverged from D. melanogaster approximately 55 and 250 million years ago, respectively.
Construction of CadN and CadN2 expression vectors
Standard PCR amplification and subcloning procedures were used to construct the expression vectors for various CadN mutants, CadN2-myc fusions, and CadN2-CadN chimera. All the cytoplasmic-domain mutants of CadN were constructed using the full-length CadN 7b-13a-18a isoform cDNA (encoding 3097 residues) as the template: CadN-Δcyto encodes 1-2943 amino-acid residues of CadN; CadN-Δβcat, 1-3018; CadN-Δjuxta; 1-2950 and 2982-3098; CadN-AAA; triple glycine-to-alanine mutations at 2966-2068. The CadN2-CadN chimera is composed of the extracellular domain (residues 1-1548) of CadN2 and the transmembrane and cytoplasmic domains (residues 2917-3098) of CadN. These cDNAs were inserted into the S2 expression vector, pRmHa3, or the P-element vector, pUAST. Constructs and detailed cloning procedures are available upon request.
Transgenic flies were generated using standard microinjection techniques in house or through Genetic Services Inc. Three or more independent transgenic lines were obtained for each construct and their CadN expression levels were assessed using the PM181-Gal4 driver and immunohistochemistry. Transgenic lines with comparable expression level (approximately 2-5 folds of the endogenous level) were used for rescue experiments.
Histology
Immunohistochemistry was performed as described previously (Ting et al., 2005). Confocal images were acquired using a Zeiss 510 META laser-scanning microscope. The obtained z-stacks of images were deconvolved using Huygens professional software (Scientific Volume Imaging) and the 3D images were rendered from the restored z-stacks using Imaris software (Bitplane Inc.)
Cell-aggregation assay
S2-suspension cells used for cell-aggregation assays were a generous gift from James Clemens. Cell culture and transfection were performed according to the Qiagen Effectene manuals. For expressing different CadN mutants in suspension S2 cells, different CadN mutant expression vectors (pRmHa3/CadN) were cotransfected with a GFP expression vector (pRmHa3/GFP). CuSO4 was added (0.7 mM) 24 hr after the transfection to induce the expression of CadN mutants and the GFP marker. The S2 cells were induced for 48 hrs and then subjected to cell-aggregation assay as described previously (Yonekura et al., 2006). The formation of cell aggregates was visualized and analyzed under a Zeiss M2bio fluorescence microscope. Cell number in each aggregate was counted using the M2bio microscope in micro mode with a 10X objective lens. The image of these cell aggregates was captured using a Zeiss Axio-Cam digital camera and processed using Openlab software. The cell aggregates were divided into five categories based on the number of cells in each aggregate (25-50, 50-100, 100-150, 150-200, and >200 cells/per aggregate). Experiments were performed in triplicate. CadN mutants were compared with the control (full-length CadN-7b-13a-18a) and confidence interval was calculated for each category.
Supplementary Material
(A) CadN CadN2 double mutant R7 axons exhibit novel phenotypes that were not observed in CadN mutants. Single CadN CadN2 double mutant R7s were generated using the GMR-Flp/MARCM system and their axonal projections (labeled with GFP [green]) were assessed at 35% APF as in Figures 3A ”-A”’. R-cell axons were visualized using MAb24B10 (red). Approximately 20% of CadN CadN2 double mutant R7 axons overshot the R7-temporary layer to reach deeper medulla layers (arrow, A, B), and some 26% exhibited expanded processes (arrows) in both R7- and R8-temporary layers (C). Inset: the red channel was removed in (C) to facilitate the visualization of growth cone processes. Scale bars: in A, 30 μm; in B, 10 μm for C.
Expressing CadN2-CadN in R7s in an otherwise wild-type background does not cause significant defects in growth cone morphology or targeting. The R7-specific driver PM181-Gal4 was used to express the mCD8GFP marker (green) (control, A,A’) or CadN2-CadN chimera (B,B’). Target selection and growth cone morphology were assessed at 17% APF. R-cell axons were visualized with MAb24B10 (red). (A’,B’) high-magnification views of (A,B). Scale bars: in A, 20 μm for B; in A’, 5 μm for B’.
LAR and Liprin-α are not required in the medulla neurons for R7 target selection. Optic lobes were rendered LAR (B, B’) or Liprin-α (C, C’) mutant with the ELF system and the mosaic tissues were marked by the loss of GFP signal (green). R7 target selection was assessed using the R-cell marker, MAb24B10 (red), in the adult animals. Removing LAR or Liprin-α in optic lobes resulted in no detectable R-cell axonal targeting defect. (A’-C’) High-magnification views of A-C, respectively. Scale bars: in A, 30 μm for B-C; in A’, 10 μm for B’-C’.
Removing CadN in medulla neurons does not cause drastic structure defects in medulla. Optic lobes were rendered wild-type (A-A’, C-C’), or CadN (B-B’,D-D’) mutant using the ELF system as in Figure 5, and the medulla neuropil structure was assessed at 17% AFP using the pan-neurite marker, Nervana (detected by anti-HRP staining, blue; A-B’) or Capricious (detected by anti-Capricious antibody, blue; C-D’). The mosaic neurons were marked by the loss of GFP signal. R-cell axons were visualized with MAb24B10 (red). Note that removing CadN in medulla neurons resulted in size reduction in the medulla and lobula neuropils. However, the layer structure in medulla appears to be largely unaffected. (A’, B’, C’, D’) High-magnification views of the medulla. ed, eye disc; la, lamina; me, medulla. Scale bars: in A for A, B, 30 μm; in A’ for B’, 10 μm; in C for C, D, 20 μm; in C’ for C’,D’, 5 μm.
Apterous positive medulla neurons express CadN. The distribution of CadN was assessed at 10% APF using anti-CadN antibody (blue). R cell axons were labeled with GMR-myr-RFP (red) and apterous positive neurons were marked with Ap-Gal4, UAS-mCD8-GFP (green). The white box in (A) demarcates the regions displayed in (A’-A”’). (A’-A”’) show the cell bodies of a group of young medulla neurons that express both apterous and CadN. CadN is enriched on the axons and processes of medulla neurons. To reveal the CadN staining in cell bodies, the signal gain was increased in (A”). An overlay of A’ and A” is shown in A”’. The apterous positive medulla neurons project axons through the medulla neuropil into the lobula. Processes extended from these axons occupy various layers in the medulla neuropil. Note that while essentially all the apterous positive neurons in the medulla express CadN, some CadN-expressing medulla neurons do not express apterous. CadN is highly expressed on R-cell axons and medulla processes. (B, B’) High-magnification views of (A). ed, eye disc; la, lamina; me, medulla: lo, lobula. Scale bars: in A, 30 μm; in B, 10 μm for B’.
Acknowledgments
We thank Iris Salecker for providing critical transgenic flies, without which this project would not be completed. We thank Tadashi Uemura for providing CadN cDNA, antibodies and transgenic flies. We thank Edward Giniger, Benjamin White, Howard Nash, Alan Hinnebusch, and Henry Levin for helpful discussion, and Margaret Dieringer for manuscript handling and editing. This work is supported by the Intramural Research Program of the NIH, National Institute of Child Health and Human Development (grant HD008748-03 to C.-H.L.) S.Y. is a fellow of the Japan Society for the Promotion of Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastasiadis PZ, Reynolds AB. Regulation of Rho GTPases by p120-catenin. Curr Opin Cell Biol. 2001;13:604–10. doi: 10.1016/s0955-0674(00)00258-1. [DOI] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Mourton T, Nixon JP, Pietz GE, Kinch M, Chen H, Brackenbury R, Rimm DL, Del Vecchio RL, Tonks NK. Dynamic interaction of PTPmu with multiple cadherins in vivo. J Cell Biol. 1998;141:287–296. doi: 10.1083/jcb.141.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga VM, Del Maschio A, Machesky L, Dejana E. Regulation of cadherin function by Rho and Rac: modulation by junction maturation and cellular context. Mol Biol Cell. 1999;10:9–22. doi: 10.1091/mbc.10.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden-Gulley SM, Payne HR, Lemmon V. Growth cones are actively influenced by substrate-bound adhesion molecules. J Neurosci. 1995;15:4370–81. doi: 10.1523/JNEUROSCI.15-06-04370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci U S A. 2006;103:11601–6. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–51. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Making connections in the fly visual system. Neuron. 2002;35:827–41. doi: 10.1016/s0896-6273(02)00876-0. [DOI] [PubMed] [Google Scholar]
- Goodwin M, Yap AS. Classical cadherin adhesion molecules: signaling and the cytoskeleton. J Mol Histol. 2004;35:839–44. doi: 10.1007/s10735-004-1833-2. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–57. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Hofmeyer K, Maurel-Zaffran C, Sink H, Treisman JE. Liprin-alpha has LAR-independent functions in R7 photoreceptor axon targeting. Proc Natl Acad Sci U S A. 2006;103:11595–600. doi: 10.1073/pnas.0604766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai Y, Hirota Y, Ozaki K, Okano H, Takeichi M, Uemura T. DN-cadherin is required for spatial arrangement ultrastructural organization of synapses. Mol Cell Neurosci. 2002;19:375–88. doi: 10.1006/mcne.2001.1081. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Kovacs EM, Ali RG, McCormack AJ, Yap AS. E-cadherin homophilic ligation directly signals through Rac and phosphatidylinositol 3-kinase to regulate adhesive contacts. J Biol Chem. 2002a;277:6708–18. doi: 10.1074/jbc.M109640200. [DOI] [PubMed] [Google Scholar]
- Kypta RM, Su H, Reichardt LF. Association between a transmembrane protein tyrosine phosphatase and the cadherin-catenin complex. J Cell Biol. 1996;139:1519–1529. doi: 10.1083/jcb.134.6.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr Biol. 2002b;12:379–82. doi: 10.1016/s0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–50. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C, Suzuki T, Gahmon G, Treisman JE, Dickson BJ. Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron. 2001;32:225–35. doi: 10.1016/s0896-6273(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The Development of Drosophila melanogaster. 2 . CSHL Press; 1993. The development of the optic lobe ; pp. 1363–1491. [Google Scholar]
- Murray MJ, Merritt DJ, Brand AH, Whitington PM. In vivo dynamics of axon pathfinding in the Drosophilia CNS: a time-lapse study of an identified motorneuron. J Neurobiol. 1998;37:607–21. [PubMed] [Google Scholar]
- Myster SH, Cavallo R, Anderson CT, Fox DT, Peifer M. Drosophila p120catenin plays a supporting role in cell adhesion but is not an essential adherens junction component. J Cell Biol. 2003;160:433–49. doi: 10.1083/jcb.200211083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–60. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Nern A, Nguyen LV, Herman T, Prakash S, Clandinin TR, Zipursky SL. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc Natl Acad Sci U S A. 2005;102:12944–9. doi: 10.1073/pnas.0502888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol. 2000;150:567–80. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TP, Bentley D. Accumulation of actin in subsets of pioneer growth cone filopodia in response to neural and epithelial guidance cues in situ. J Cell Biol. 1993;123:935–48. doi: 10.1083/jcb.123.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–26. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- Owens DW, McLean GW, Wyke AW, Paraskeva C, Parkinson EK, Frame MC, Brunton VG. The catalytic activity of the Src family kinases is required to disrupt cadherin-dependent cell-cell contacts. Mol Biol Cell. 2000;11:51–64. doi: 10.1091/mbc.11.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacquelet A, Lin L, Rorth P. Binding site for p120/delta-catenin is not required for Drosophila E-cadherin function in vivo. J Cell Biol. 2003;160:313–9. doi: 10.1083/jcb.200207160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne HR, Burden SM, Lemmon V. Modulation of growth cone morphology by substrate-bound adhesion molecules. Cell Motil Cytoskeleton. 1992;21:65–73. doi: 10.1002/cm.970210108. [DOI] [PubMed] [Google Scholar]
- Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat Neurosci. 2005;8:443–50. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler S, Suzuki E, Chiba A. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat Neurosci. 2000;3:1012–7. doi: 10.1038/79833. [DOI] [PubMed] [Google Scholar]
- Roura S, Miravet S, Piedra J, Garcia de Herreros A, Dunach M. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J Biol Chem. 1999;274:36734–40. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–30. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Shinza-Kameda M, Takasu E, Sakurai K, Hayashi S, Nose A. Regulation of layer-specific targeting by reciprocal expression of a cell adhesion molecule, capricious. Neuron. 2006;49:205–13. doi: 10.1016/j.neuron.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–5. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Takeichi M, Nakagawa S. Identification of a nonchordate-type classic cadherin in vertebrates: chicken Hz-cadherin is expressed in horizontal cells of the neural retina and contains a nonchordate-specific domain complex. Dev Dyn. 2004;229:899–906. doi: 10.1002/dvdy.10493. [DOI] [PubMed] [Google Scholar]
- Tayler TD, Garrity PA. Axon targeting in the Drosophila visual system. Curr Opin Neurobiol. 2003;13:90–5. doi: 10.1016/s0959-4388(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Ting CY, Lee CH. Visual circuit development in Drosophila. Curr Opin Neurobiol. 2007;17:1–8. doi: 10.1016/j.conb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, Lee CH. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–63. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- Yap AS, Kovacs EM. Direct cadherin-activated cell signaling: a view from the plasma membrane. J Cell Biol. 2003;160:11–6. doi: 10.1083/jcb.200208156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura S, Ting CY, Neves G, Hung K, Hsu SN, Chiba A, Chess A, Lee CH. The variable transmembrane domain of Drosophila N-cadherin regulates adhesive activity. Mol Cell Biol. 2006;26:6598–608. doi: 10.1128/MCB.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) CadN CadN2 double mutant R7 axons exhibit novel phenotypes that were not observed in CadN mutants. Single CadN CadN2 double mutant R7s were generated using the GMR-Flp/MARCM system and their axonal projections (labeled with GFP [green]) were assessed at 35% APF as in Figures 3A ”-A”’. R-cell axons were visualized using MAb24B10 (red). Approximately 20% of CadN CadN2 double mutant R7 axons overshot the R7-temporary layer to reach deeper medulla layers (arrow, A, B), and some 26% exhibited expanded processes (arrows) in both R7- and R8-temporary layers (C). Inset: the red channel was removed in (C) to facilitate the visualization of growth cone processes. Scale bars: in A, 30 μm; in B, 10 μm for C.
Expressing CadN2-CadN in R7s in an otherwise wild-type background does not cause significant defects in growth cone morphology or targeting. The R7-specific driver PM181-Gal4 was used to express the mCD8GFP marker (green) (control, A,A’) or CadN2-CadN chimera (B,B’). Target selection and growth cone morphology were assessed at 17% APF. R-cell axons were visualized with MAb24B10 (red). (A’,B’) high-magnification views of (A,B). Scale bars: in A, 20 μm for B; in A’, 5 μm for B’.
LAR and Liprin-α are not required in the medulla neurons for R7 target selection. Optic lobes were rendered LAR (B, B’) or Liprin-α (C, C’) mutant with the ELF system and the mosaic tissues were marked by the loss of GFP signal (green). R7 target selection was assessed using the R-cell marker, MAb24B10 (red), in the adult animals. Removing LAR or Liprin-α in optic lobes resulted in no detectable R-cell axonal targeting defect. (A’-C’) High-magnification views of A-C, respectively. Scale bars: in A, 30 μm for B-C; in A’, 10 μm for B’-C’.
Removing CadN in medulla neurons does not cause drastic structure defects in medulla. Optic lobes were rendered wild-type (A-A’, C-C’), or CadN (B-B’,D-D’) mutant using the ELF system as in Figure 5, and the medulla neuropil structure was assessed at 17% AFP using the pan-neurite marker, Nervana (detected by anti-HRP staining, blue; A-B’) or Capricious (detected by anti-Capricious antibody, blue; C-D’). The mosaic neurons were marked by the loss of GFP signal. R-cell axons were visualized with MAb24B10 (red). Note that removing CadN in medulla neurons resulted in size reduction in the medulla and lobula neuropils. However, the layer structure in medulla appears to be largely unaffected. (A’, B’, C’, D’) High-magnification views of the medulla. ed, eye disc; la, lamina; me, medulla. Scale bars: in A for A, B, 30 μm; in A’ for B’, 10 μm; in C for C, D, 20 μm; in C’ for C’,D’, 5 μm.
Apterous positive medulla neurons express CadN. The distribution of CadN was assessed at 10% APF using anti-CadN antibody (blue). R cell axons were labeled with GMR-myr-RFP (red) and apterous positive neurons were marked with Ap-Gal4, UAS-mCD8-GFP (green). The white box in (A) demarcates the regions displayed in (A’-A”’). (A’-A”’) show the cell bodies of a group of young medulla neurons that express both apterous and CadN. CadN is enriched on the axons and processes of medulla neurons. To reveal the CadN staining in cell bodies, the signal gain was increased in (A”). An overlay of A’ and A” is shown in A”’. The apterous positive medulla neurons project axons through the medulla neuropil into the lobula. Processes extended from these axons occupy various layers in the medulla neuropil. Note that while essentially all the apterous positive neurons in the medulla express CadN, some CadN-expressing medulla neurons do not express apterous. CadN is highly expressed on R-cell axons and medulla processes. (B, B’) High-magnification views of (A). ed, eye disc; la, lamina; me, medulla: lo, lobula. Scale bars: in A, 30 μm; in B, 10 μm for B’.


