Abstract
Covalent modifications of histone N-terminal tails are required for the proper assembly and activation of the general transcription factors at promoters. Here, we analyze histone acetylation and phosphorylation in Drosophila transgenes activated by the yeast Gal4 transcriptional activator in the context of different promoters. We show that, independent of the promoter, transcription does not correlate with acetylation of either H3-Lys 14 or H4-Lys 8. Histone H3 associated with the DNA of Gal4-induced transcribing transgenes driven by the Drosophila Hsp70 promoter is hyperphosphorylated at Ser 10 during transcription. Surprisingly, histone H3 at Gal4-induced transgenes driven by the P element Transposase promoter is not hyperphosphorylated. The data suggest that transcription occurs without acetylated H4 and H3 in both transgenes in Drosophila polytene chromosomes. Instead, phosphorylation of H3 is linked to transcription and can be modulated by the structure of the promoter.
Keywords: Histone, phosphorylation, acetylation, transcription, chromatin
Posttranslational covalent modifications of N-terminal tails of the core histones at the nucleosome play an important role in determining chromatin structure, which is an essential component of the control of nuclear biology (Jenuwein and Allis 2001). The expression of a large number of genes from organisms across the phylogenetic scale correlates with histone modifications such as acetylation, methylation, phosphorylation, and ubiquitination in the nucleosomes surrounding the promoter of the gene (Roth et al. 2001; Berger 2002; Fry and Peterson 2002; Sun and Allis 2002). Acetylation of histones H3 and H4 N-terminal tails has been broadly associated with activation of transcription. Histone acetyl-transferases (HATs) are found in a variety of coactivators and transcription factor complexes, whereas histone deacetylases (HDACs) are present in protein complexes with a repressive function (Roth et al. 2001). H3 methylation at Lys 9 has the opposite effect and therefore is found in regions where transcription is repressed by chromatin structure (Rea et al. 2000; Bannister et al. 2001). Phosphorylation of H3 at Ser 10 has also been correlated with activation of transcription in yeast, mammals, and Drosophila (Cheung et al. 2000; Lo et al. 2000; Nowak and Corces 2000; Lo et al. 2001; Thomson et al. 2001; Li et al. 2002; Strelkov and Davie 2002).
Two nonexclusive models have been postulated to explain the role of histone N-terminal tail modification in transcription. The first one suggests that histone modifications have an effect on the relative charge of the histone tails, rendering a more open or closed chromatin state that determines the accessibility of transcription factors to the core promoter. The second model, termed the histone code hypothesis, predicts that a combination of covalent modifications of histone tails functions as a target for the specific binding of effector proteins (Turner 2000; Jenuwein and Allis 2001). Binding of these proteins will ultimately determine the transcriptional state of the chromatin. The histone code hypothesis is gaining support following the realization that well characterized proteins with functions involving chromatin structure and control of gene expression contain domains that selectively bind covalently modified histone N-tails. For example, proteins containing a bromodomain specifically bind lysine-acetylated histone tails. Bromodomain proteins include many transcriptional regulators such as several HATs and important components of the transcription machinery such as TAF250 (Jacobson et al. 2000). In addition, proteins containing a chromodomain specifically bind methylated histone tails. For example, HP1 is a chromodomain-containing protein involved in the formation of heterochromatin and in the silencing of gene expression by binding methylated H3 at Lys 9 (Bannister et al. 2001).
It is not clear whether H3 phosphorylation at Ser 10 also functions as a recognition site for specific binding of chromatin-associated proteins. In vivo and in vitro data suggest that acetylation of H3 at Lys 14 and phosphorylation at Ser 10 are coupled and that H3 acetylation by yeast GCN5 is enhanced by phosphorylation at Ser 10 (Cheung et al. 2000; Lo et al. 2000). This association correlates with transcription activation and is supported as well by structural data showing a specific interaction between arginine 164 of GCN5 and the phosphorylated Ser 10 of the H3 N-terminal tail. Other results, however, question the general implication of this model when applied to other organisms; for example, the dramatic changes in H3 phosphorylation that take place during the heat shock response in Drosophila are not followed by equivalent changes in histone acetylation (Nowak and Corces 2000). The histone H3 N-terminal tails at the Drosophila heat shock genes become hyperphosphorylated at Ser 10 upon induction of transcription by heat shock. Ser 10-phosphorylated H3 is also observed under normal conditions at many other sites in polytene chromosomes, including actively transcribing regions such as ecdysone-induced puffs. In addition, phosphorylated histone H3 disappears from all nonheat shock loci during the heat shock response, coinciding with the shutdown of transcription of most genes, further supporting the hypothesis that H3 phosphorylation may have a central role in the control of gene expression in Drosophila. Except for a weak acetylation signal in some of the heat shock puffs, the global changes observed in H3 phosphorylation after heat shock are not followed by detectable equivalent changes in H3 or H4 acetylation (Nowak and Corces 2000).
To gain further insights into the role of H3 phosphorylation during transcription in Drosophila and its correlation with acetylation, we addressed the question of whether phosphorylation and acetylation at specific residues of the histone N-terminal tails are associated with transcription of transgenes ectopically activated by the Gal4 transcriptional activator. In yeast, the acidic domains of the widely used exogenous transcriptional activators Gal4 and VP16 recruit the SAGA complex to the promoter of the gene (Bhaumik and Green 2001; Larschan and Winston 2001). Because complexes equivalent to SAGA are also found in humans and in Drosophila, where they contain PCAF, a protein homologous to the GCN5 present in the yeast's SAGA (Aoyagi and Wassarman 2000), we asked whether transcriptional activation by Gal4 in Drosophila involves the same type of covalent histone tail modifications as in yeast. The results suggest that phosphorylation and not acetylation of H3 accompanies transcriptional activation of these transgenes, and this modification is not determined by the nature of the transcriptional activator but rather by the structure of the core promoter itself.
Results and Discussion
Gal4-induced transcription of Hsp70 promoter-driven transgenes is associated with H3 hyperphosphorylation at Ser 10
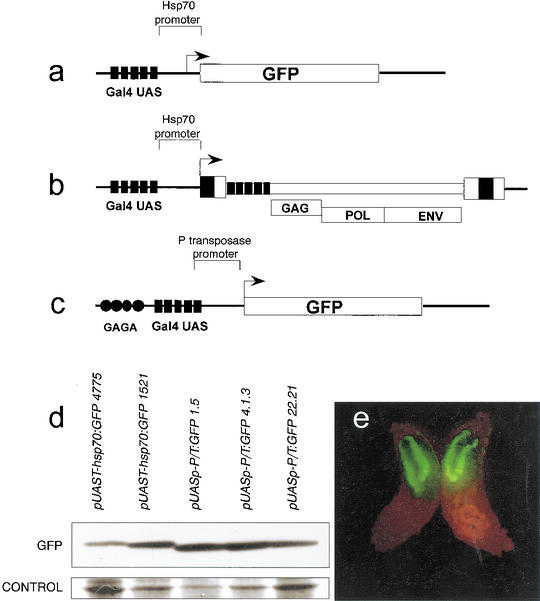
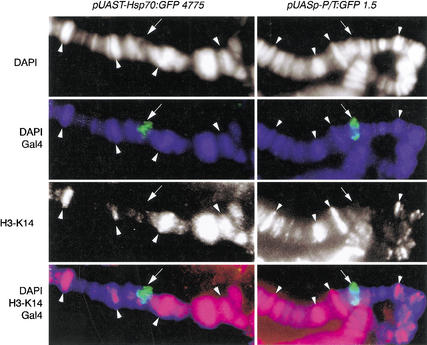
Immunostaining using specific antibodies has shown that a large number of sites in Drosophila polytene chromosomes contain phosphorylated histone H3 at Ser 10. The same experiments show that activation of transcription by the heat shock factor (HSF) at the heat shock genes induces high levels of H3 phosphorylation at the site of transcription (Nowak and Corces 2000). We first addressed the question of whether H3 phosphorylation at the Hsp70 promoter depends exclusively on HSF or can take place by activation of transcription mediated by any other transcription factor. A simple way to address this question is to analyze histone H3 phosphorylation at nucleosomes linked to the DNA of transcribing transgenes that use the TATA box promoter of the Hsp70 gene but lack the HSF binding sites. We thus tested whether transcription of a GFP reporter gene under the control of Gal4–UAS sites fused to the promoter region of the Hsp70 gene was associated with H3 phosphorylation. To this end we crossed flies transformed with the pUAST–Hsp70:GFP transgene (Fig. 1a) with flies carrying a transgene encoding Gal4 under the control of the Hsp70 promoter (Hsp70:Gal4). This Gal4 transgene is constitutively expressed in salivary glands, and therefore no heat shock was necessary to induce expression of Gal4. Transcription of the GFP gene induced by Gal4 was monitored by Western blot analysis and by observing the salivary glands under a dissecting fluorescence microscope (Fig. 1d,e). Only larvae carrying both transgenes (pUAST–Hsp70:GFP and Hsp70:Gal4) show GFP fluorescence (e.g., see Fig. 1e); the levels of GFP expression were quantitated more precisely by Western analysis of GFP protein produced in larvae carrying each transgene (Fig. 1d). Antibodies specific against phosphorylated H3 at Ser 10 were then used to perform immunostaining experiments on polytene chromosome spreads. In order to determine whether the pUAST–Hsp70:GFP transgene colocalized with phosphorylated H3 histones, the same spreads were coimmunostained using monoclonal antibodies specific against the DNA binding domain of the Gal4 protein, which should label only the locus occupied by the transgene containing Gal4–UAS DNA sequences. Results show that, upon activation of transcription by Gal4, phosphorylated H3 histones are found in the pUAST–Hsp70:GFP transgene (Fig. 2). These results suggest that the Hsp70 promoter plus an exogenous transcriptional activator are sufficient to induce H3 phosphorylation at Ser 10 during transcription, indicating that HSF is not unique in the induction of H3 phosphorylation. In addition, the results indicate the presence of phosphorylated H3 in most polytene chromosome interbands and not in bands. We also found an almost perfect correlation between the presence of phosphorylated H3 at Ser 10 and the GAGA protein (data not shown). Since GAGA is a general transcriptional activator (Granok et al. 1995) and strong evidence indicates that transcription in polytene chromosomes occurs in interbands and not in bands, where chromatin is highly compacted (Sass 1982; Weeks et al. 1993), these observations suggest that histone phosphorylation is a landmark for actively transcribing regions in Drosophila polytene chromosomes.
Figure 1.
Structure of reporter genes used for analysis of histone modifications and expression analysis. (a) pUAST–Hsp70:GFP. (b) pUAST–Hsp70:gypsy. (c) pUASp–P/T:GFP. (d) Western analysis of GFP expression in pUAST–Hsp70:GFP and pUASp–P/T:GFP in Hsp70:Gal4 larvae. Numbers in the names of the transgenes indicate different independent lines. Loading controls correspond to Actin proteins as determined by molecular weight after staining of the blotted membrane with Coomassie blue. (e) An example of GFP fluorescence in salivary glands in larvae expressing a pUAST–Hsp70:GFP transgene.
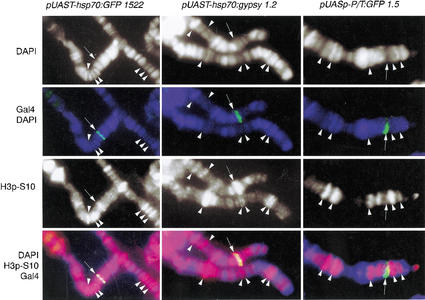
Figure 2.
Correlation between histone H3 phosphorylated at Ser 10 and expression of various transgenes. Phosphorylated H3-Ser 10 is shown in red, Gal4 in green, and DAPI in blue. Arrows point to the localization of each transgene. Arrowheads show colocalization of polytene chromosome interbands (no DAPI staining) and phosphorylated H3.
We then asked whether the recruitment of factors controlling H3 phosphorylation depends on the structure of the promoter. The Hsp70 promoter contains a consensus TATA box, plus an Initiator and downstream sequences that may influence the transcriptional activity of the gene (Lee et al. 1992; Wu et al. 2001). To test whether downstream sequences have a role in H3 phosphorylation, we fused the origin of transcription containing the Initiator and downstream sequences of the gypsy retrotransposon (Arkhipova et al. 1986), to the upstream promoter region of Hsp70 containing the TATA box (Fig. 1b). The pUAST–Hsp70:gypsy transgene contains the same TATA box from Hsp70 as pUAST–Hsp70:GFP but differs from this transgene in the sequences downstream of the original transcription. These sequences correspond to those of the gypsy retrovirus and include regions R and U5 of the LTR plus binding sites for the Suppressor of Hairy wing protein [Su(Hw)] and all sequences encoding GAG, POL, and ENV proteins (Fig. 1b). Immunostaining experiments using antibodies raised against the gypsy Envelope protein (ENV) show that gypsy is actively expressed on salivary glands from larvae carrying the pUAST–Hsp70:gypsy and the pUAST:Gal4 transgenes, whereas no ENV protein was found in larvae not expressing Gal4 (data not shown). This result indicates that gypsy is actually transcribed from the pUAST–Hsp70:gypsy transgene and not from gypsy copies elsewhere in the genome. Immunolocalization experiments to detect phosphorylated H3 were performed with pUAST–Hsp70:gypsy, and results were identical to those obtained with the pUAS–Hsp70:GFP transgene. H3 is always heavily phosphorylated at the site where pUAST–Hsp70:gypsy and the Gal4 protein are present (Fig. 2). The conclusion of this experiment is that phosphorylation of H3 at Ser 10 at the Hsp70 promoter is not dependent on specific sequences located downstream of the initiation of transcription.
Phosphorylation of H3 might require binding of TBP at the core promoter
To further investigate the effect of promoter structure on histone H3 phosphorylation, we analyzed a promoter completely unrelated to the Hsp70 promoter. We found a candidate in the P Transposase promoter (P/T) present in the pUASp transformation vector developed by P. Rorth (1998). The transcription start site of the P Transposase promoter has been determined experimentally to be nucleotide 87 of the P element sequence, with the putative TATA box (tatacact) found at −28 (O'Hare and Rubin 1983). The presence of a C at position 5 of the TATA box is highly unusual and has not been observed in any of the 900 characterized promoters described in the eukaryotic promoter database (EPD). In addition, it has been shown experimentally that the presence of a C at this position will disrupt the interaction with the TATA binding protein (TBP; Patikoglou et al. 1999; Praz et al. 2002). The P Transposase promoter also lacks Initiator and DPE elements. These structural characteristics might explain the ability of the P transposase promoter to drive transcription in the germ line, whereas the Hsp70 promoter is not expressed in these cells (Rorth 1998). We constructed a reporter gene containing the promoter region of the Drosophila P element Transposase fused to the coding region of the GFP gene (pUASp–P/T:GFP; Fig. 1c). Surprisingly, when we analyzed histone modifications on transcribing pUASp–P/T:GFP transgenes, we failed to detect H3 phosphorylation at the sites of insertion (Fig. 2). Because these results could be attributed to position effects associated with a particular transgene, we repeated the same experiments with two additional pUASp–P/T:GFP and pUAST–Hsp70:GFP transgenes inserted in different positions in the genome (see Materials and Methods). In all additional lines, transcription of pUAST–Hsp70:GFP transgenes also correlated with hyperphosphorylation of H3, whereas chromatin associated with all pUASp–P/T:GFP transgenes showed nondetectable H3 phosphorylation (data for all additional lines not shown). Because higher levels of H3 phosphorylation may correlate only with high levels of transcription, it is possible that the differences in the level of H3 phosphorylation observed between Hsp70 and P transposase promoters were in fact due to differences in promoter strength. To test this possibility, we performed a Western blot analysis comparing GFP levels in third-instar larvae carrying pUASp–P/T:GFP or pUAST–Hsp70:GFP transgenes in the presence of Gal4 expressed from the same Hsp70:Gal4 transgene. Results show that the amount of GFP does not correlate with the amount of H3 phosphorylation observed, suggesting that differences in H3 phosphorylation between promoters are due to factors other than the strength with which promoters activate transcription (Fig. 1d).
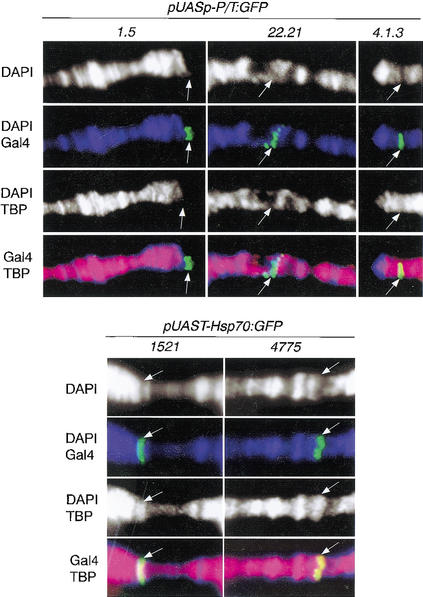
The Hsp70 promoter contains a consensus TATA box that binds TBP. Because TFIID complexes can be assembled without the participation of TBP (Verrijzer 2001), we asked whether the P Transposase promoter actually recruits TBP. To this end, we performed coimmunolocalization experiments using antibodies against TBP and Gal4 in both pUASp–P/T:GFP and pUAST–Hsp70:GFP transgenes. The results suggest that TBP binds to the Hsp70 promoter but is missing from the P Transposase promoter (Fig. 3), implying that the assembly of general transcription factors may follow different biochemical pathways in terms of chromatin structure, depending on specific signals at the core promoter. In addition, promoter-specific differences in H3 phosphorylation also suggest that the kinase responsible for such phosphorylation might be recruited by components associated with general transcription factors at the core promoter and is independent of the nature of the transcriptional activator. The factor that recruits such a kinase must be directly dependent on the presence of the TBP protein. In Drosophila, TRF1 and TRF2 are two known general transcription factors found in salivary glands that can replace the function of TBP in transcription initiation at certain promoters (Hansen et al. 1997; Rabenstein et al. 1999; Freiman et al. 2001). It follows from our data that the assembly of the preinitiation complex by different components of TFIID may also determine changes in the chromatin structure at the trancribing gene. Finding these components, in particular the kinase responsible for the phosphorylation of histone H3, might be an important key to better understand the complexity of transcription regulation in higher eukaryotes.
Figure 3.
Analysis of TBP localization in various transgenes. TBP staining is shown in red, DAPI in blue, and Gal4 in green. Arrows show the position of the transgene in different stocks.
Transcription of Gal4-activated transgenes is independent of acetylation of histone H4 at Lys 8 and histone H3 at Lys 14
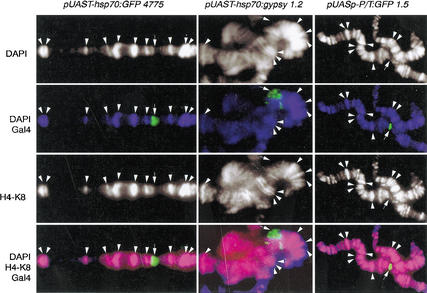
Because the genomic distribution of acetylated histones is apparently unaltered after the dramatic changes in gene transcription that take place during the heat shock response (Nowak and Corces 2000), we decided to determine whether transcription correlates with acetylation of specific histones in the transgenes analyzed above. We first used antibodies specific against acetylated histone H4 at Lys 8, which has been correlated with actively transcribing chromatin (Strahl and Allis 2000). Immunostaining of polytene chromosome spreads utilizing the same procedures as those described above were performed. Sites of acetylated H4 at Lys 8 do not colocalize with Gal4 labeling, which corresponds to the sites of the actively transcribing GFP transgenes in all of the promoters assayed (Fig. 4).
Figure 4.
Acetylated H4 at Lys 8 does not colocalize with actively transcribed transgenes. Acetylated H4 at Lys 8 is shown in red, Gal4 in green, and DAPI in blue. Arrows show localization of each transgene. Arrowheads show colocalization of polytene chromosome bands and acetylated H4 at Lys 8.
Because Gal4 probably recruits the SAGA complex to the promoters of the transgenes, we decided to also test whether there is a functional link in Drosophila between H3 phosphorylation and H3 acetylation, as occurs in yeast (Lo et al. 2000). Analysis of the amino acid sequence of GCN5 from distantly related organisms such as yeast, Drosophila, and humans shows a striking conservation of the arginine at position 164, suggesting that this protein has retained its ability to recognize phosphorylated H3-Ser 10 across the phylogenetic scale. In Xenopus oocytes, for example, the activation of transcription by the thyroid hormone receptor also correlates with an increase of H3 phosphorylated at Ser 10 and acetylated at Lys 14, suggesting that acetylation and phosphorylation are also coupled during transcription activation in higher eukaryotes (Li et al. 2002). If transcriptional activation by Gal4 in Drosophila also involves the recruitment of the SAGA complex to the promoter, the Drosophila GCN5 (dPCAF) protein may have a role in the histone modifications undergone by the GFP transgenes used in the present study. We therefore performed immunostaining experiments using antibodies specific against acetylated H3 at Lys 14, the amino acid target for the yeast acetyl transferase GCN5. Because pUAST–Hsp70:GFP transgenes are hyperphosphorylated and pUASp–P/T:GFP transgenes are not, one can argue that dPCAF might be involved in the activation of transcription of the former, through acetylation of H3 at Lys 14, but not in the activation of transcription of the latter. Surprisingly, H3 is not acetylated at detectable levels in any of the two transgenes (Fig. 5). These results suggest that acetylation of H4 at Lys 8 or H3 at Lys 14 is not required for activation of transcription in either of the transgenes used in this study. Whether dPCAF is recruited by Gal4 to the promoter of the transgenes cannot be determined with the data presented here. However, even if dPCAF is recruited to the promoter of the Gal4–UAS transgenes, we can conclude that the function of H3 phosphorylation observed here is independent of a requirement for phosphorylation of Ser 10 as a prerequisite to enhance acetylation of Lys 14.
Figure 5.
Acetylated H3 at Lys 14 does not colocalize with transcribing transgenes or with polytene chromosome interbands. Acetylated H3 at Lys 14 is shown in red, Gal4 in green, and DAPI in blue. Arrows show insertion sites of the transgene. Arrowheads show colocalization of acetylated H3 at Lys 14 only with polytene chromosome bands.
In support of this observation, we found that H3 phosphorylated at Ser 10 is always localized in interbands (see Fig. 2), whereas acetylated H4 and H3 always localize in the bands (Figs. 4, 5). Because transcription in polytene chromosomes localizes mostly to the interbands, these results further support the idea that histone phosphorylation, and not acetylation, is widely associated with active transcription in Drosophila. In addition to the distribution of acetylated histones H3 at Lys 14 and H4 at Lys 8 described in this work, identical polytene band–interband distribution has been found for polyacetylated H3, acetylated H4-Lys 5, and acetylated H4-Lys 12 (Pile and Wassarman 2000). In light of the large amount of evidence suggesting a role for histone acetylation in transcription activation, Pile and Wassarman (2000) interpreted these results as suggesting that immunolocalization on polytene chromosomes might not be sufficiently sensitive to detect changes in histone acetylation after transcription initiation, and that the pattern of histone acetylation along the chromosome might be a reflection of background acetylation, only visible in polytene bands where chromatin is highly compacted. Evidence against this possibility comes from the observation that acetylated H3 at Lys 14 (Fig. 5) and polyacetylated H3 (data not shown) are not present in all bands, whereas acetylated H4 at Lys 8 is present in almost all polytene bands (Fig. 4). This unequal distribution indicates that, at least for some acetylated histones, the polytene chromosome distribution reflects differences in chromatin organization and not only in chromatin compaction. This observation suggests that the lack of histone acetylation at sites of active transcription is not a technical artifact and might, in fact, be a reflection of the absence of acetylated histones in these regions. Alternatively, the lack of acetylated histones at sites of Gal4-induced transcription could indicate that histone acetylation is only transient during the initiation of transcription and is not required at subsequent steps.
The discrepancy between results in yeast and vertebrates and in Drosophila argues in favor of two separate roles for H3 phosphorylation. One role might correspond to the observed general association of transcription with H3 phosphorylation in polytene chromosomes, which seems to be independent of acetylation. In support of this observation, histone phoshporylation and histone acetylation are independently regulated during the activation of transcription of c-fos and c-jun in mouse (Thomson et al. 2001). The second role might be related to the conservation of arginine 164 in GCN5, which would serve as a recognition site for the binding of PCAF and subsequent acetylation of Lys 14, as occurs in yeast and in genes activated by the thyroid hormone receptor. An interesting possibility for a role of H3 phosphorylation is suggested by recent reports indicating that the histone H3.3 variant replaces H3 in regions of active transcription in Drosophila (Ahmad and Henikoff 2002a,b). It is possible that the H3 phosphorylation we observe is mechanistically linked to the replacement of H3 in Drosophila, and therefore that the same mechanism is not employed by yeast, which lacks histone H3 (Ahmad and Henikoff 2002b).
Materials and methods
Transgenes and transgenic flies
pUAST–Hsp70:GFP transgenes were obtained from the Bloomington Drosophila stock center. The stock numbers used in this work were 4775 (chromosome 2), 1521 (chromosome 2), 1522 (chromosome 3), and 5193 (chromosome 1). In order to obtain the pUAST–Hsp70:gypsy transgene, the PUAST transformation vector, containing the core promoter of the Hsp70 gene (Brand and Perrimon 1993), was used to clone the origin of transcription of gypsy by inserting a DNA fragment, starting at the nucleotide 234 position of the retrovirus, into the origin of transcription of the Hsp70 promoter. This construct removes the initiator element and all downstream sequences from the Hsp70 promoter and leaves intact the Initiator element from gypsy and the sequences upstream of the Hsp70 transcription initiation site in pUAST. In the second transgene, pUASp–P/T:GFP, the GFP DNA sequence was inserted into the NotI site of the pUASp vector (Rorth 1998). Each plasmid was microinjected into y w; P(Δ2-3)/TM6 embryos, and flies carrying the transgene inserted in the TM6 (pUAST–Hsp70:GFP1.5) chromosome were selected by standard procedures (Robertson et al. 1988).To obtain new insertion sites, both Drosophila stocks were crossed to y w; P(Δ2-3)/TM6. New insertions in the X (pUAST–Hsp70:GFP4.1.3) and in the second chromosome (pUAST–Hsp70:GFP 22.21) were recovered. TheHsp70:Gal4 transgene was obtained from Dr. Allen Shearn (Department of Biology, Johns Hopkins University, Baltimore, MD).
Immunocytochemistry and Western blot analysis
Western analysis was carried out by standard procedures using SuperSignal West Pico Chemiluminescent substrate from Pierce for detection. Protein extracts were prepared from equal amounts of larvae carrying different transgenes expressing GFP in their salivary glands and loaded in the gels. After immunodetection, membranes were stained with Coomassie blue to further control for loading of the samples. Immunolocalization of proteins on polytene chromosomes was as described (Harrison et al. 1993). Anti-Ser 10 phosphohistone H3 antibodies were obtained from Dr. David Allis (Department of Biochemistry and Molecular Genetics, University of Virginia H.S.C., Charlottesville, VA) and Upstate Biotechnology, and anti-Lys 8 acetyl H4 and anti-Lys 14 acetyl H3 antibodies were obtained from Upstate Biotechnology. Anti TBP antibodies were provided by Dr. James T. Kadonaga (Section of Molecular Biology, University of California, San Diego). Anti-Gal4 DBD monoclonal antibody was obtained from Santa Cruz Biotechnology. Antibodies against gypsy Envelope protein were previously obtained in our laboratory (Song et al. 1997). Proteins were visualized using FITC- or Texas red-conjugated secondary antibodies (Jackson Immunoresearch Laboratories); DNA was stained with DAPI and chromosomes were examined using a Zeiss Axiophot microscope and a Photometrics cooled CCD camera (Roper Scientific).
Acknowledgments
We thank Drs. F. Mongelard and S. Nowak for valuable discussions and suggestions, P. Plata-Rengifo for the generation of the pUASp–P/T:GFP construct, and Drs. J.T. Kadonaga and D. Allis for kindly providing antibodies against Drosophila TBP and Ser 10 phosphohistone H3, respectively. This work was supported by U.S. Public Health Service Award GM35463 from the N.I.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL corces@jhu.edu; FAX (410) 516-5456.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1021403.
References
- Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci. 2002a;12:12. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002b;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Aoyagi N, Wassarman DA. Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: Diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation. J Cell Biol. 2000;150:45–50. doi: 10.1083/jcb.150.2.f45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova IR, Mazo AM, Cherkasova VA, Gorelova TV, Schuppe NG, Llyin YV. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986;44:555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- Fry CJ, Peterson CL. Transcription. Unlocking the gates to gene expression. Science. 2002;295:1847–1848. doi: 10.1126/science.1070260. [DOI] [PubMed] [Google Scholar]
- Granok H, Leibovitch BA, Shaffer CD, Elgin SC. Chromatin. Ga-ga over GAGA factor. Curr Biol. 1995;5:238–241. doi: 10.1016/s0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- Hansen SK, Takada S, Jacobson RH, Lis JT, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- Harrison DA, Gdula DA, Coyne RS, Corces VG. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes & Dev. 1993;7:1966–1978. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes & Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Li J, Lin Q, Yoon HG, Huang ZQ, Strahl BD, Allis CD, Wong J. Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor. Mol Cell Biol. 2002;22:5688–5697. doi: 10.1128/MCB.22.16.5688-5697.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Lo W-S, Duggan L, Tolga Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1—A histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes & Dev. 2000;14:3003–3013. doi: 10.1101/gad.848800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K, Rubin GM. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes & Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pile LA, Wassarman DA. Chromosomal localization links the SIN3–RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 2000;19:6131–6140. doi: 10.1093/emboj/19.22.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praz V, Perier R, Bonnard C, Bucher P. The eukaryotic promoter database, EPD: New entry types and links to gene expression data. Nucleic Acids Res. 2002;30:322–324. doi: 10.1093/nar/30.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein MD, Zhou S, Lis JT, Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Sass H. RNA polymerase B in polytene chromosomes: Immunofluorescent and autoradiographic analysis during stimulated and repressed RNA synthesis. Cell. 1982;28:269–278. doi: 10.1016/0092-8674(82)90345-2. [DOI] [PubMed] [Google Scholar]
- Song SU, Kurkulos M, Boeke JD, Corces VG. Infection of the germ line by retroviral particles produced in the follicle cells: A possible mechanism for the mobilization of the gypsy retroelement of Drosophila. Development. 1997;124:2789–2798. doi: 10.1242/dev.124.14.2789. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strelkov IS, Davie JR. Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res. 2002;62:75–78. [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Mahadevan LC. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol Cell. 2001;8:1231–1241. doi: 10.1016/s1097-2765(01)00404-x. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP. Transcription factor IID—Not so basal after all. Science. 2001;293:2010–2011. doi: 10.1126/science.1064980. [DOI] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: Correlations with gene activity and transcript processing. Genes & Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Wu CH, Madabusi L, Nishioka H, Emanuel P, Sypes M, Arkhipova I, Gilmour DS. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol Cell Biol. 2001;21:1593–1602. doi: 10.1128/MCB.21.5.1593-1602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]