Abstract
We show that the Xenopus homologs of Ndc80/Tid3/HEC1 (xNdc80) and Nuf2/MPP1/Him-10 (xNuf2) proteins physically interact in a 190-kD complex that associates with the outer kinetochore from prometaphase through anaphase. Injecting function-blocking antibodies to either xNdc80 or xNuf2 into XTC cells caused premature exit from mitosis without detectable chromosome congression or anaphase movements. Injected cells did not arrest in response to microtubule drugs, showing that the complex is required for the spindle checkpoint. Kinetochores assembled in Xenopus extracts after immunodepletion of the complex did not contain xRod, xZw10, xP150 glued (Dynactin), xMad1, xMad2, xBub1, and xBub3, demonstrating that the xNdc80 complex is required for functional kinetochore assembly. In contrast, function-blocking antibodies did not affect the localization of other kinetochore proteins when added to extracts containing previously assembled kinetochores. These extracts with intact kinetochores were deficient in checkpoint signaling, suggesting that the Ndc80 complex participates in the spindle checkpoint. We also demonstrate that the spindle checkpoint can arrest budding yeast cells lacking Ndc80 or Nuf2, whereas yeast lacking both proteins fail to arrest in mitosis. Systematic deletion of yeast kinetochore genes suggests that the Ndc80 complex has a unique role in spindle checkpoint signaling. We propose that the Ndc80 complex has conserved roles in kinetochore assembly, chromosome congression, and spindle checkpoint signaling.
Keywords: Cell cycle, centromere, kinetochore, chromosome segregation, mitosis, spindle checkpoint
Kinetochores are large, multiprotein structures that are assembled on centromeric chromatin and have at least three important functions to coordinate chromosome segregation: bind spindle microtubules, coordinate chromosome movements, and activate the spindle checkpoint. The fully assembled kinetochore consists of >20 proteins (Choo 1997). It has an inner core including the Cenp-A and Cenp-C proteins, which localize to the centromere throughout the cell cycle and are thought to organize and anchor the kinetochore to centromeric heterochromatin (Earnshaw and Rothfield 1985; Saitoh et al. 1992). The inner centromere protein (Incenp) and the kinesin XKCM1 (MCAK) interact with the centromeric chromatin between sister kinetochores specifically in mitosis (Cooke et al. 1987; Wordeman et al. 1991). Kinetochores also have outer domains that contain numerous proteins including Cenp-F, Rod, Zw10, the homologs of yeast mitotic spindle checkpoint proteins, and the microtubule-interacting proteins Cenp-E, dynein, and dynactin (a dynein regulator; Choo 1997). Many outer kinetochore proteins are assembled into the kinetochore after nuclear envelope breakdown, and subgroups leave the kinetochore at three points of mitosis: in response to microtubule capture, during early anaphase, and at the end of anaphase (Choo 1997; Shah and Cleveland 2000).
To ensure that every sister-chromatid pair achieves proper bipolar-microtubule attachment before anaphase onset, cells have evolved a regulatory mechanism referred to as the spindle checkpoint (Gardner and Burke 2000; Shah and Cleveland 2000). Proteins that mediate the spindle checkpoint were originally identified in Saccharomyces and are encoded by BUB1, BUB2, BUB3, MAD1, MAD2, MAD3, MPS1, and BFA1 (Hoyt et al. 1991; Li and Murray 1991; Winey et al. 1991; Pereira et al. 2000; Wang et al. 2000). Vertebrate homologs of most of these checkpoint proteins localize to kinetochores and are required for spindle checkpoint arrest (Shah and Cleveland 2000). The residence times of GFP–Mad2 and GFP–Cdc20 at unattached kinetochores in cultured cells have half-lives of 24–28 and 6 sec, respectively, suggesting that checkpoint proteins can rapidly cycle through unattached kinetochores (Howell et al. 2000; Kallio et al. 2002). This rapid cycling can explain how a Mad2/Cdc20 complex that is generated at unattached kinetochores disperses throughout the cell and inhibits anaphase.
A recently identified complex of four proteins, Ndc80/Tid3 (hereafter referred to as Ndc80), Nuf2, Spc24, and Spc25, has been purified from yeast cells and localized to the kinetochore (Janke et al. 2001; Wigge and Kilmartin 2001). All four proteins have been shown to be required for microtubule binding in yeast, as has the Nuf2 homolog in Caenorhabditis elegans and Ndc80 in human cells (Durfee et al. 1993; Chen et al. 1997; Wigge et al. 1998; Howe et al. 2001; Janke et al. 2001; Nabetani et al. 2001). Two of the proteins in this complex (hereafter referred to as the Ndc80 complex) have been implicated in the spindle checkpoint. Temperature-sensitive spc24 and spc25 mutants cannot arrest in mitosis, whereas most ndc80 and nuf2 mutants do arrest (Osborne et al. 1994; Wigge et al. 1998; He et al. 2001; Janke et al. 2001; Wigge and Kilmartin 2001). Homologs of Ndc80 and Nuf2 exist from fission yeast to humans, yet Spc24 and Spc25 homologs have not been identified in metazoans. Thus, Ndc80 and Nuf2 have a conserved role in microtubule binding; however, it is unclear whether they are also required for kinetochore assembly or spindle checkpoint signaling.
We have cloned and characterized the Xenopus homologs of Ndc80 (xNdc80) and Nuf2 (xNuf2). The xNdc80 and xNuf2 proteins interact biochemically and localize to mitotic kinetochores from prometaphase through anaphase. The xNdc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint function in both tissue culture cells and in Xenopus eggs extracts. Yeast cells lacking either Ndc80 or Nuf2 are checkpoint-proficient; however, cells lacking both proteins are checkpoint-deficient. To determine if checkpoint-deficient phenotypes are common for yeast mutants lacking kinetochore proteins, we performed a systematic analysis of yeast mutants lacking kinetochore proteins. Only a small number of presently identified kinetochore proteins participate in the spindle checkpoint. We propose that the Ndc80 complex is required for normal kinetochore function in chromosome congression and plays a pivotal role in coordinating kinetochore activity with the spindle checkpoint.
Results
Identification of Xenopus Ndc80 and Nuf2
We cloned the Xenopus homologs of Ndc80 and Nuf2 from a stage-11.5–14 cDNA library to investigate if the Ndc80 complex has a conserved role in the vertebrate kinetochore. The full-length xNuf2 sequence encodes a 462-amino-acid protein with a predicted molecular mass of 54.4 kD, and the full-length xNdc80 sequence encodes a 638-amino-acid protein with a predicted molecular mass of 74.5 kD. The xNdc80 and xNuf2 proteins share 72% and 63% similarity with their human counterparts, and both have 40% similarity to their yeast homologs (Supplemental Fig. 1A,B). There are C-terminal coiled-coil domains from amino acids 143–277 and 311–462 of xNuf2 and from 249–394 and 479–638 of xNdc80 (Supplemental Fig. 1C; Lupas et al. 1991). Similar coiled-coil regions are present in all homologs, and yeast two-hybrid analysis suggests that the two proteins directly interact via these domains (Janke et al. 2001; Nabetani et al. 2001; Wigge and Kilmartin 2001). The N-terminal portions of both proteins have no identifiable domains.
Affinity-purified xNuf2 antibody recognizes a specific protein at 50 kD in both interphase egg extracts and XTC tissue culture cell lysates. Affinity-purified xNdc80 antibody recognizes a doublet protein at 75 kD in interphase egg extract and a single 74-kD band in XTC cell lysate (Fig. 1A). Lambda phosphatase treatment did not alter the gel mobility of the xNdc80 protein, which suggests that the protein is not phosphorylated (data not shown). By comparing the immunoblot signal of Xenopus interphase extract with known amounts of recombinant protein, we estimate that the concentration of xNuf2 is 53 nM and that of xNdc80 is 66 nM in Xenopus egg extracts (data not shown).
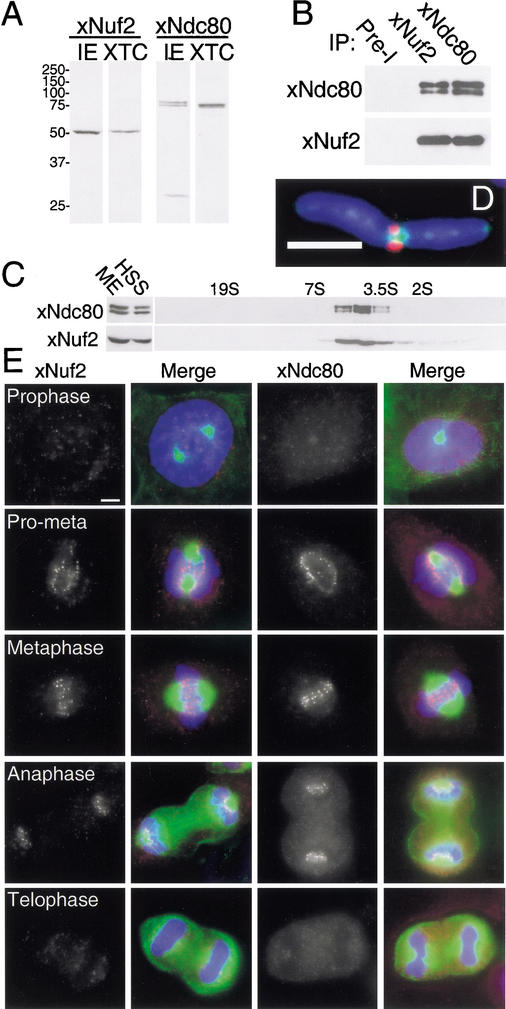
Figure 1.
xNdc80 and xNuf2 form a complex in vivo that localizes to kinetochores from prometaphase through anaphase. (A) Characterization of polyclonal antibodies generated against xNuf2 and xNdc80. Affinity-purified anti-xNuf2 antibodies recognize a single 50-kD protein in both Xenopus interphase egg extract (IE) and XTC cell lysate (XTC). Affinity-purified anti-xNdc80 antibodies recognize a doublet protein at 75 kD in Xenopus interphase egg extract and a single protein at 75 kD in XTC cell lysate. The small band identified in the xNdc80 lane is either a degradation product or a cross-reacting protein. (B) Coimmunoprecipitation of xNdc80 and xNuf2 from Xenopus egg extracts. xNdc80 and xNuf2 protein were immunoprecipitated (IP) from interphase extract and blotted for xNuf2 and xNdc80. Preimmune (Pre-I) sera controls did not precipitate either xNuf2 or xNdc80 protein. (C) xNdc80 and xNuf2 cosediment on a sucrose density gradient as a 4.3S complex. High-speed supernatants (HSS) of mitotic extracts (ME) were sedimented through 5%–30% sucrose density gradient and blotted for xNdc80 and xNuf2. (D) Localization of xNdc80 on a mitotic chromosome. XTC cells, arrested in mitosis with nocodazole, were fixed and stained with anti-xINCENP (green), biotinylated anti-xNdc80 antibody (red), and Hoeschst 33342 to visualize DNA (blue). (E) xNdc80 and xNuf2 are associated with the kinetochore from prometaphase through anaphase. Cycling XTC cells were fixed and stained with mouse anti-tubulin antibody (green) and either anti-xNuf2 or anti-xNdc80 antibody (red). DNA visualized with Hoechst 33342 (blue). Bars: D,E, 5 μm.
xNdc80 and xNuf2 physically associate in vivo
To determine if the physical interaction between Ndc80 and Nuf2 is conserved in vertebrates, we immunoprecipitated both xNdc80 and xNuf2 from Xenopus egg extracts and immunoblotted the resulting precipitates. xNdc80 protein was detected in xNuf2 immunoprecipitates and xNuf2 was detected in xNdc80 immunoprecipitates, demonstrating that these two proteins interact (Fig. 1B). The association is not cell-cycle-regulated in the early embryo (data not shown). Immunodepletion of xNdc80 also removed xNuf2, which demonstrates that the majority of xNuf2 is physically associated with xNdc80 (see Fig. 4B, below). Immunodepletion with anti-xNuf2 antibodies did not fully deplete either xNdc80 or xNuf2. Preimmune sera controls did not precipitate either xNdc80 or xNuf2 protein.
Figure 4.
xNdc80 and xNuf2 are required for spindle checkpoint signaling in Xenopus egg extracts and localization of xMad1, xMad2, xBub1, xBub3, xRod, xZw10, and xP150 glued (dynactin) to the kinetochore. (A) xNdc80 and xNuf2 are required for spindle checkpoint establishment in Xenopus extracts. CSF-arrested extracts were preincubated for 30 min with control IgG, anti-xNuf2, or anti-xNdc80 antibodies. Sperm nuclei and nocodazole were added for 30 min, followed by the addition of calcium chloride. Samples were taken before (t = 0) the addition of calcium and every 15 min thereafter to monitor histone H1 kinase activity. Autoradiograms of histone H1 kinase assays as well as photographs of nuclear morphology (t = 60) are shown. (B) Anti-xNdc80 antibodies deplete both xNdc80 and xNuf2 proteins. An immunoblot of 1 μL each of Xenopus egg extract depleted with preimmune sera (Pre-I Δ) or anti-xNdc80 (xNdc80 Δ) antibodies was probed with anti-xNdc80 and anti-xNuf2 antibodies. (C) Depletion of the xNdc80 complex inhibits xMad1, xMad2, xBub1, xBub3, xRod, xZw10, and xP150 glued (dynactin) localization to kinetochores. CSF-arrested extracts were preimmune-sera-depleted or xNdc80-depleted, and sperm and nocodazole were added. Following 30 min, sperm chromatin was spun onto coverslips and fixed. Kinetochores were stained for xNdc80, xNuf2, xCenp-A, XKCM1, xP150 glued (dynactin), xRod, xZw10, xMad1, xMad2, xBub1, or xBub3 (red). Chromatin was visualized with Hoechst 33342 (blue). Representative pictures of xNdc80, xRod, xP150 glued, and xCenp-A are shown. (D) Quantification of chromatin with positive kinetochore staining. One hundred nuclei from C were scored for positive kinetochore staining of xNdc80, xNuf2, xCenp-A, XKCM1, xP150 glued (dynactin), xRod, xZw10, xMad1, xMad2, xBub1, or xBub3 from preimmune-sera- or xNdc80-depleted extracts. Black and gray bars represent preimmune sera and xNdc80 depletions, respectively. Bars: A,C, 5 μm.
Xenopus extracts were clarified by centrifugation, and both xNdc80 and xNuf2 were found in the high-speed supernatant, suggesting that they exist in a soluble complex (Fig. 1C). High-speed supernatants were fractionated on a Superose 6 column (Supplemental Fig. 2) and sedimented through 5%–30% sucrose gradients; the resulting fractions were immunoblotted for xNdc80 and xNuf2 (Fig. 1C; Supplemental Fig. 2B). The majority of both proteins comigrated on both gel filtration and sucrose gradient. The size appeared identical in both interphase and mitotic egg extracts. xNdc80 and xNuf2 fractionated at ∼1 MD on gel filtration and sedimented as a 4.3S complex on a sucrose gradient. We determined that xNdc80 and xNuf2 exist in a 190-kD complex (Siegel and Monty 1966). The complex is highly extended, reminiscent of long coiled-coil proteins like myosin.
Localization of xNdc80 and xNuf2 in mitosis
To determine if xNdc80 and xNuf2 localize to the kinetochore, XTC cells were arrested with nocodazole and stained with anti-xNdc80 antibody. Kinetochores were identified by costaining for xINCENP, a passenger protein that localizes to the inner centromere (Cooke et al. 1987). xNdc80 localized to two dots at the primary constriction of condensed chromosomes, and xINCENP staining was in the middle of the two dots (Fig. 1D; Supplemental Fig. 2C). The Ndc80 complex does not require microtubules to associate with kinetochores. xNdc80 colocalized with xNuf2 at the kinetochore (Supplemental Fig. 2D).
Analysis of asynchronously growing XTC cells showed little nuclear staining in interphase or prophase; however, both xNdc80 and xNuf2 associated with kinetochores in prometaphase (Fig. 1E; data not shown). Both proteins were detected at kinetochores of aligning chromosomes at prometaphase and remained until telophase. This localization is consistent with the previously published observations and is consistent with the proposed microtubule-binding function of Ndc80 and Nuf2 in yeast, nematodes, and human tissue culture cells (Osborne et al. 1994; Chen et al. 1997; Wigge et al. 1998; Howe et al. 2001; Martin-Lluesma et al. 2002).
XTC cells injected with function-blocking anti-xNdc80 or anti-xNuf2 antibodies undergo cytokinesis and mitotic exit without chromosome alignment and segregation
To determine the function of xNdc80 and xNuf2, we injected control IgG, anti-xNdc80, or anti-xNuf2 antibodies into prophase or prometaphase XTC cells. Cytokinesis was induced within 20 minutes after anti-xNdc80 injection (n = 8) or anti-xNuf2 injection (n = 6). The cytokinetic furrows cleaved through the chromosomes that had not separated, reminiscent of a “cut” phenotype observed in some mitotic mutants of Schizosaccharomyces pombe (Fig. 2A; Supplemental Videos 1, 2). Cells injected with either anti-xNdc80 or anti-xNuf2 antibodies entered interphase ∼45 min after injection without anaphase chromosome movements. Control injected cells progressed normally through mitosis and entered interphase ∼62 min following injection (Supplemental Video 3). Anti-xNdc80 and anti-xNuf2 antibody injections induce premature cytokinesis and exit from mitosis in the absence of anaphase chromosome separation and movement.
Figure 2.
Injection of anti-xNdc80 and anti-xNuf2 antibodies into mitotic XTC cells induces a “cut” phenotype, defects in chromosome alignment, and diminution of kinetochore fiber microtubule bundles. (A) An XTC cell was injected with anti-xNuf2 antibody at early prometaphase, incubated for ∼60 min, fixed, immunolabeled to visualize the injected antibody, and stained with DAPI to visualize DNA. Cell cleavage resulted in a “cut” phenotype with unequally sized daughter nuclei connected by a chromatin bridge (arrow). (B) XTC cells injected with anti-xNuf2 antibody at early mitosis were incubated for 2 h in the presence of MG132, fixed, immunolabeled to visualize the injected antibody, and stained with DAPI. Many of the antibody-injected cells contained several unaligned chromosomes (arrowheads), whereas the majority of noninjected, mitotic cells (yellow arrow) in the same population had arrested with all chromosomes aligned at the metaphase plate. The inset shows a higher magnification of an anti-xNuf2-antibody-injected cell to reveal that the injected antibody concentrates at presumptive kinetochores. (C) Microinjection of anti-xNdc80 antibody also blocked chromosome alignment in cells maintained in the M phase with MG132. Labeling with anti-tubulin antibody reveals changes in spindle structure compared with an uninjected control cell on the same coverslip. Note the decrease in organized bundles of kinetochore microtubules extending from the spindle poles to the chromosomes. The inset shows the injected antibody labeled with secondary anti-rabbit IgG. Bar, 10 μm.
Normal mitotic exit requires degradation of cell cycle regulators by the 26S proteasome (King et al. 1996). To determine if the induced mitotic exit was proteasome-dependent, we injected cells cultured in the presence of the proteasome inhibitor MG132. Mitotic exit was blocked for >6 h after injecting either anti-xNdc80 or anti-xNuf2 antibodies (data not shown). Exit from mitosis induced by antibody injection is proteasome-dependent.
Cells exposed to MG132 arrest at metaphase with their chromosomes aligned at the spindle equator. MG132 treatment blocked mitotic exit in XTC cells injected with anti-xNdc80 and anti-xNuf2 antibody; therefore, we could assess the effects of injecting antibodies on chromosome alignment, independent of the effects on cytokinesis and mitotic exit. Mitotic cells injected with anti-xNdc80 or anti-xNuf2 antibody in the presence of MG132 failed to align chromosomes at the metaphase plate, unlike nearby uninjected cells (Fig. 2B,C). Chromosomes failed to fully congress in 86% (19 out of 22) of cells injected with anti-xNdc80 antibody and 48% (12 out of 25) of cells injected with anti-xNuf2 antibody, in the presence of MG132. These results are consistent with previous reports of anti-HEC1 (Ndc80 homolog) antibody injection in mammalian cells and RNAi-mediated depletion of HIM-10 (Nuf2 homolog) in C. elegans embryos (Chen et al. 1997; Howe et al. 2001).
Cells were injected with anti-xNdc80, anti-xNuf2, or control IgG antibodies in the presence of MG132, to determine whether the defects in chromosome congression were accompanied by alterations in the organization of the mitotic spindle. Chromosome congression was inhibited, and chromosomes were often randomly distributed over the mitotic spindle (Fig. 2C). Kinetochore fiber bundles of microtubules were evident from the spindle poles to the chromosomes in uninjected cells. Kinetochore fiber bundles of microtubules were severely reduced in the majority of cells (22 of 30) injected with anti-xNdc80, suggesting the attachment or stabilization of microtubules with kinetochores was impaired (Fig. 2C). Similar results were seen in anti-xNuf2-injected cells.
The lack of complete chromosome congression only partially explains the “cut” phenotype seen after antibody injection into non-drug-treated cells (Fig. 2A). Normally, the presence of unaligned chromosomes activates the spindle checkpoint, and cells are blocked from exiting the M phase. The premature exit from mitosis in the presence of misaligned chromosomes induced by anti-xNdc80 and anti-xNuf2 antibodies suggested that the spindle checkpoint was abrogated in antibody-injected cells. To determine if the Ndc80 complex was required for spindle checkpoint signaling, XTC cells were incubated with taxol, injected in prophase with anti-xNdc80 or anti-xNuf2 antibodies, and subjected to real-time imaging. Injected cells exited from mitosis and reentered interphase ∼80 min after nuclear envelope breakdown as visualized by surface membrane blebbing, which typically accompanies the onset of anaphase, and by subsequent reformation of the nuclear envelope (Supplemental Video 4). Uninjected or control-injected XTC cells remained arrested for >6 h. Cells injected in the presence of both MG132 and taxol did not exit from mitosis for at least 6 h, demonstrating that mitotic exit was proteasome-dependent (data not shown).
Taxol-treated XTC cells were injected with either anti-xNdc80 or anti-xNuf2 antibodies and incubated for 120 min. Cells were fixed and stained for DNA and with fluorescent anti-rabbit IgG to identify injected cells; 90% of the cells injected with either antibody exited mitosis in the presence of taxol (Fig. 3). There were two obvious phenotypes. Either cytokinetic furrows failed resulting in polyploid cells containing multiple misshapen nuclei, or chromatin bridges connected two daughter nuclei through the midbody. Identical results were obtained when injected XTC cells were treated with nocodazole (data not shown). Microinjection of either anti-xNdc80 or anti-xNuf2 antibody into XTC cells abrogates the spindle checkpoint.
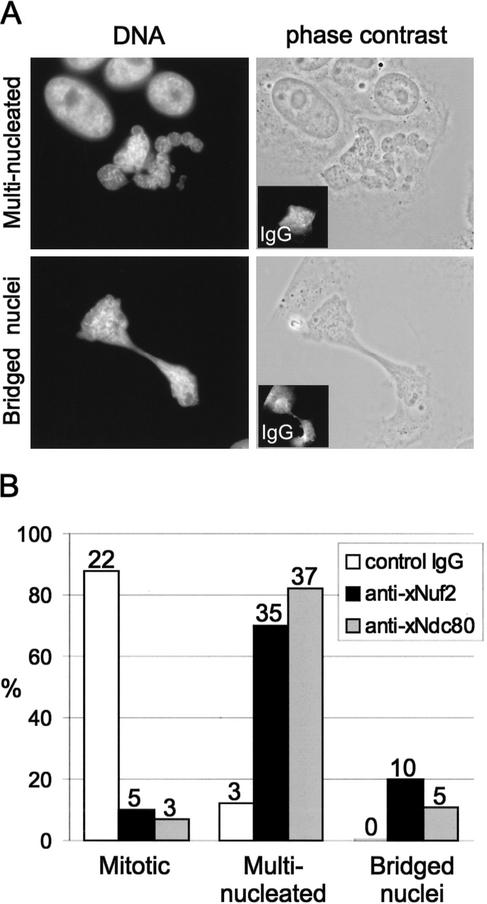
Figure 3.
Anti-xNdc80- and anti-xNuf2-antibody-injected cells bypass the spindle checkpoint. Mitotic XTC cells, treated with taxol, were injected with either anti-xNdc80 or anti-xNuf2 antibodies. After an additional 2 h, the cells were fixed, immunolabeled with fluorescent anti-rabbit IgG secondary to locate injected cells, and stained with DAPI. (A) Examples of the two major phenotypes resulting from anti-xNuf2 antibody injection in taxol. There were either multinucleated polyploid cells or cells with a “cut” phenotype. The insets show the injected antibody detected by fluorescent anti-rabbit IgG secondary. (B) Quantification of injected cells. The numbers above the bars indicate the number of injected cells in each category.
The Ndc80 complex is required for the spindle checkpoint in Xenopus egg extracts
We used Xenopus cycling extracts that can be induced for spindle checkpoint activity (Minshull et al. 1994) to investigate the role of xNdc80 and xNuf2 in the spindle checkpoint. CSF-extracts containing sperm, nocodazole, and control IgG established a spindle checkpoint and remained arrested in mitosis for at least 60 min (Fig. 4A). Control CSF-extracts without nocodazole did not arrest. In contrast, extracts preincubated with either anti-xNdc80 or anti-xNuf2 antibodies prior to the addition of sperm and nocodazole failed to arrest in mitosis and entered interphase because they exhibited low histone H1 kinase activity and interphase nuclear morphology. Both xNdc80 and xNuf2 are required to establish the spindle checkpoint in Xenopus extracts.
The xNdc80 complex is required for xP150 glued, xRod, xZw10, Mad1, Mad2, Bub1, and Bub3 to assemble on kinetochores
We determined whether xNdc80 and xNuf2 are required to localize the kinetochore proteins xCENP-A, XKCM1, xRod, xZw10, xP150 glued (Dynactin), xMad1, xMad2, xBub1, and xBub3 to kinetochores. Both xNdc80 and xNuf2 were depleted from Xenopus CSF-extracts using anti-xNdc80 antibodies. Western blot analysis showed that depletion of xNdc80 removed >95% of xNdc80 and all detectable xNuf2 protein from the extract (Fig. 4B). Sperm and nocodazole were added to depleted extracts, and sperm chromatin was spun onto coverslips and processed for immunofluorescence. The outer kinetochore proteins xNdc80, xNuf2, xP150 glued (Dynactin), xRod, xZw10, xMad1, xMad2, xBub1, and xBub3 were not detected at most kinetochores in xNdc80-depleted extracts (Fig. 4C,D), whereas the inner kinetochore proteins xCenp-A and XKCM1 showed positive kinetochore staining. All tested kinetochores proteins were easily detected in preimmune serum-depleted extracts. Thus association of a number of outer kinetochore proteins requires the Ndc80 complex. The loss of checkpoint activity after the addition of xNdc80 and xNuf2 antibodies can be explained by the inability to assemble checkpoint proteins at kinetochores.
Adding function-blocking antibodies to Xenopus extracts containing preassembled kinetochores abrogates checkpoint signaling without dramatically affecting kinetochore structure
To separate the role of the Ndc80 complex in kinetochore assembly from a role in the spindle checkpoint, we added either control IgG or anti-xNdc80 antibodies to Xenopus extracts containing preassembled kinetochores in nocodazole. Extracts that received control IgG antibodies remained in mitosis, demonstrating that the extracts were competent in spindle checkpoint signaling (Fig. 5A). However, extracts that received anti-xNdc80 antibodies exited mitosis, suggesting that they were defective in the spindle checkpoint. The Ndc80 complex is required to maintain the spindle checkpoint.
Figure 5.
Addition of anti-xNdc80 antibodies to preassembled kinetochores disrupts spindle checkpoint signaling, but does not dramatically affect kinetochore assembly. (A) Sperm nuclei and nocodazole were incubated with CSF-arrested extracts for 30 min to assemble kinetochores and establish the spindle checkpoint. Control IgG or anti-xNdc80 antibody was then added for an additional 30 min before release from CSF arrest with calcium. Samples were taken before (t = 0) the addition of calcium and every 15 min thereafter to monitor histone H1 kinase activity. Autoradiograms of histone H1 kinase assays as well as photographs of nuclear morphology (t = 60) are shown. (B) Kinetochore structure is not severely disturbed following addition of anti-xNdc80 antibodies to a Xenopus extract containing preassembled kinetochores. Sperm nuclei and nocodazole were incubated with CSF-arrested extracts for 30 min. Control IgG or anti-xNdc80 antibody was then added for 30 min. Sperm chromatin was subsequently spun onto coverslips, fixed, and processed for immunofluorescence. Chromatin was stained for xNdc80 (by adding a CY-3-conjugated sheep anti-rabbit secondary antibody, “2° only”), xCenp-A, xP150 glued (Dynactin), or xRod (red). Chromatin was visualized with Hoechst 33342 (blue). Bars: A,B, 5 μm.
We localized four kinetochore proteins to determine if adding the function-blocking xNdc80 antibodies affected the structure of the outer kinetochore. xNdc80 remained on kinetochores. The staining intensity of the inner kinetochore protein xCenpA and the outer kinetochore proteins xP150 glued (Dynactin), xRod, and xZw10 were unchanged by anti-xNdc80 antibody addition (Fig. 5B, xZw10; data not shown). The addition of function-blocking anti-xNdc80 antibodies to preassembled kinetochores abrogated spindle checkpoint signaling without severely affecting kinetochore structure.
ndc80-1 and nuf2-61 mutants arrest at metaphase by the spindle checkpoint
Budding yeast mutants of the Ndc80 complex exhibit strikingly different phenotypes. Although all mutants cannot bind microtubules, loss of either Spc24 or Spc25 results in chromosome missegregation and generation of cells with 4C DNA content. In contrast, loss of either Ndc80 or Nuf2 can arrest cells in mitosis with a 2C DNA content (Janke et al. 2001; Wigge and Kilmartin 2001). One interpretation is that kinetochores are detached from the spindle in all the mutants but that spc24-1 and spc25-1 are checkpoint-defective and ndc80-1 and nuf2-61 cells are checkpoint-proficient. To confirm this, wild-type, ndc80-1, and nuf2-61 cells were synchronized with α-factor at 23°C, incubated at 37°C to inactivate the proteins, and released into the cell cycle. We measured DNA content, Pds1 levels, and cellular morphology (Fig. 6A,B). The ndc80-1 and nuf2-61 cells accumulated predominantly with a 2C DNA content, whereas wild-type cells continued to divide. A small percentage of the nuf2-61 cells appeared with a 1C DNA content after prolonged incubation at the restrictive temperature (210 min), suggesting that some of the cells continued into the next cell cycle.
Figure 6.
ndc80-1 and nuf2-61 arrest at metaphase in an MAD2- and BUB2-dependent manner. Cells were synchronized in G1 with α-factor and released into the cell cycle at the restrictive temperature (37°C). Cells were assayed for DNA content, budding index, and Pds1 levels. Cycling cells (cyc) were also examined for Pds1 levels. Tubulin (Tub2) serves as a loading control. Wild-type (WT) cells (A), ndc80-1 and nuf2-61 mutants (B), and ndc80-1 and nuf2-61 mutants (C) containing deletions of either MAD2 or BUB2. Squares represent small budded cells, triangles represent large budded cells, and circles represent multibudded cells.
To determine if the mutants arrested at metaphase, we measured the stability of Pds1, which is degraded as cells enter anaphase (Fig. 6A,B). Wild-type cells entered anaphase by degrading Pds1 by 60 min. In contrast, Pds1 levels remained high in ndc80-1 and nuf2-61 mutants, indicating they arrested at metaphase. There was a decrease in Pds1 levels in the nuf2-61 cells (180 min) that precedes the accumulation of cells with a 1C content of DNA. The nuf2-61 mutant is partially functional at the restrictive temperature, and given sufficient time, a small percentage of cells escape the arrest.
To determine if this metaphase arrest in ndc80-1 and nuf2-61 is dependent on the spindle checkpoint, we deleted either MAD2 or BUB2 in these mutants. The double mutants could not arrest and accumulated with a greater than 2C DNA content (Fig. 6C). In addition, a substantial proportion of multibudded cells accumulated, suggesting they had proceeded to the next cell cycle. These results show that, unlike spc24-1 and spc25-1 cells, the ndc80-1 and nuf2-61 mutants were arrested at metaphase by the spindle checkpoint. We were surprised that the arrest was affected by bub2 because it is thought to inhibit mitosis in response to spindle misorientation. Perhaps Ndc80 and Nuf2 have additional roles in regulating spindle positioning.
The spindle checkpoint is activated in degron-tagged alleles of Ndc80 or Nuf2
We were concerned that the nuf2-61 and ndc80-1 mutations retained partial function. We therefore constructed heat-inducible “degron-tagged” alleles of both genes (Gardner et al. 2001). We assayed synchronized ndc80dt and nuf2dt cells containing degron fusion proteins for checkpoint proficiency by flow cytometry. Cells were arrested with α-factor under permissive conditions, the degron-tagged proteins were eliminated, and cells were then released and assayed by flow cytometry. Ndc80 and Nuf2 fusion protein levels were significantly reduced 60 min following release from α-factor (Fig. 7A). Both ndc80dt and nuf2dt cells arrested with a 2C DNA content (Fig. 7A). Few cells with a 1C DNA content accumulated in the nuf2dt mutant, suggesting that it is a more severe allele than nuf2-61. Eliminating Ndc80 and Nuf2 function by two complementary approaches has generated cells that are proficient for spindle checkpoint function.
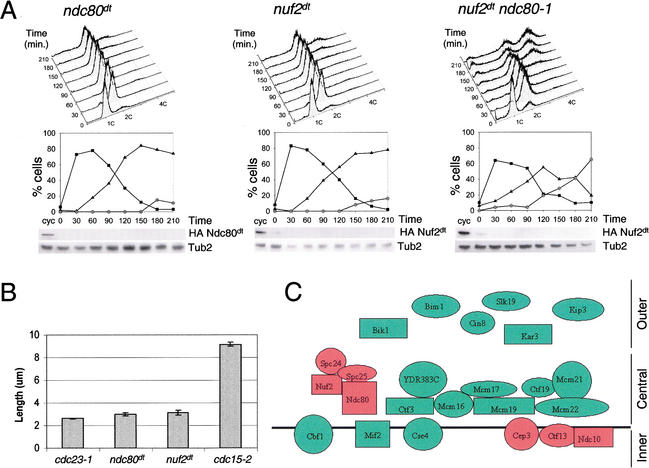
Figure 7.
Characterization of degron-tagged ndc80 and nuf2 alleles. (A) ndc80dt and nuf2dt mutants. Cells were synchronized in G1 with α-factor and released into the cell cycle under degron-restrictive conditions. Cells were assayed for DNA content, budding index, and degron-tagged protein levels. Tubulin (Tub2) serves as a loading control. Squares represent small budded cells, triangles represent large budded cells, and circles represent multi-budded cells. (B) ndc80dt and nuf2dt cells arrest with short, broken, and often misaligned spindles that are slightly longer than the metaphase spindles of cdc23-1 cells, but distinct from the long anaphase spindles of cdc15-2 cells. Spindles were measured from ndc80dt and nuf2dt cells fixed at 120 min after release from α-factor into the cell cycle under degron-inducing conditions. Spindle lengths were measured in cdc23-1 (n = 68), cdc15-1 (n = 69), ndc80dt (n = 101), and nuf2dt (n = 91) cells. Error bars indicate standard error. (C) Systematic deletion of yeast kinetochore genes. Six deletion mutants lacking central kinetochore proteins and four mutants lacking outer kinetochore proteins were analyzed for checkpoint proficiency 6 h following nocodazole treatment. A summary of the data from these experiments and those described in Gardner et al. (2001) are shown. Red shapes indicate those proteins required for spindle checkpoint activity, and green shapes indicate proteins dispensable for checkpoint function. The black line represents centromeric DNA. Inner, central, and outer kinetochore designations are defined as described (Cheeseman et al. 2002). Shapes that are touching have been shown to interact biochemically.
ndc80dt and nuf2dt mutants show divided nuclei and aberrant spindles
We determined the spindle and nuclear morphology of metaphase-arrested ndc80dt and nuf2dt cells. Approximately 75% of both mutants arrested with a single DNA mass, whereas 25% accumulated with two asymmetrically divided nuclei. The cells often contained aberrant or fractured spindles, in which tubulin staining was reduced in the spindle midzone (data not shown). The asymmetric DNA staining that we found in the ndc80dt and nuf2dt mutants appears identical to the previous characterization of ndc80-1 and nuf2-61 cells (Osborne et al. 1994; Wigge et al. 1998). Other kinetochore mutants are similar, for example, mif2-1, spc34-3 dam1-1, and ask1-1 arrest with a 2C DNA and have fractured spindles with partially divided nuclei (Brown et al. 1993; Jones et al. 1999; Li et al. 2002).
To confirm that the cells arrested in metaphase, we measured the length of the spindle in ndc80dt and nuf2dt mutants and compared it to spindles in a cdc23-1 mutant (metaphase) and a cdc15-2 mutant (anaphase). The distance between spindle poles in both ndc80dt and nuf2dt mutants was slightly longer than the cdc23-1 spindle length and substantially shorter than the elongated spindles of anaphase-arrested cdc15-2 cells (Fig. 7). The metaphase spindle length is consistent with the conclusions that ndc80dt and nuf2dt have defective kinetochore attachment and arrest in metaphase.
Ndc80 and Nuf2 have a redundant checkpoint role
To determine if Ndc80 and Nuf2 have redundant roles in the checkpoint, we constructed a strain that lacks both proteins. Because the ndc80-1 nuf2-61 double mutant was synthetically lethal (Wigge and Kilmartin 2001), we constructed an ndc80-1 nuf2dt strain. The cells could not arrest at metaphase and rereplicated their DNA in the subsequent cell cycle as assayed by flow cytometry. The ndc80-1 nuf2dt double mutant could not arrest in mitosis (Fig. 7A) and therefore eliminating both proteins abrogates the spindle checkpoint in Xenopus and in yeast.
One trivial explanation is that any mutant that eliminates kinetochore function will inactivate the spindle checkpoint. We previously analyzed kinetochore proteins for checkpoint proficiency and extended the analysis to other proteins of the kinetochore (Gardner et al. 2001). We analyzed six deletion mutants lacking central kinetochore proteins (Ctf3, Mcm16, Mcm17, Mcm19, Mcm 22, and YDR383C, a component of the Mcm19 complex) and five mutants lacking outer kinetochore proteins (Bik1, Bim1, Cin8, Slk19, and Kip3) for the ability to arrest in response to nocodazole. All deletion mutants of nonessential kinetochore genes were checkpoint-proficient (Supplemental Fig. 3B; data not shown). A summary of the data from these experiments and from Gardner et al. (2001) is presented in Figure 7C. Checkpoint function, analyzed in these mutants, maps to only two identified kinetochore complexes, CBF3 and Ndc80. Moreover, it is unlikely that the checkpoint phenotypes of the Ndc80 complex are caused by the loss of the tested central and outer kinetochore proteins.
Discussion
Ndc80 and Nuf2 are components of a recently described complex of evolutionarily conserved proteins that are important for kinetochore assembly and function (He et al. 2001; Janke et al. 2001; Wigge and Kilmartin 2001). The Xenopus homologs of Ndc80 and Nuf2 are physically associated in a 190-kD complex and localize to kinetochores in XTC cells from prometaphase through anaphase. Injection of function-blocking antibodies against either xNdc80 or xNuf2 into XTC cells induced premature mitotic exit and inhibited chromosome congression. Injected cells also failed to arrest in response to microtubule poisons, suggesting a role in spindle checkpoint signaling. In egg extracts immunodepleted of the xNdc80 complex, several outer kinetochore proteins failed to assemble. We also added anti-xNdc80 antibodies to extracts containing preassembled kinetochores and found conditions that retained the association of outer kinetochore proteins, including the spindle checkpoint proteins xRod and xZw10 (Basto et al. 2000; Chan et al. 2000). Under these conditions, extracts were defective in spindle checkpoint activity. We performed a systematic analysis of kinetochore mutants to identify kinetochore proteins required for the spindle checkpoint in budding yeast. Besides the previously examined CBF3 complex, members of the Ndc80 complex were the only kinetochore proteins that had phenotypes consistent with roles in checkpoint signaling. In contrast to our findings in Xenopus, both ndc80-1 and nuf2-61 mutants in budding yeast arrest in mitosis in response to the spindle checkpoint. We addressed this difference between Xenopus and yeast regarding the functions of Ndc80 and Nuf2 and showed that eliminating both proteins in yeast abrogated the spindle checkpoint. The effect cannot be caused by removing most central and outer kinetochore proteins, because our systematic analysis indicates that each mutant is checkpoint-proficient. We propose that the Ndc80 complex has multiple functions during mitosis and is directly involved in spindle checkpoint function in organisms as diverse as yeast and Xenopus.
The kinetochore is an integral component of the spindle checkpoint, and cells arrest in the cell cycle when chromosomes are misaligned (Shah and Cleveland 2000). Present models suggest that the kinetochore initiates the inhibitory signal. A molecular understanding of how either lack of tension or microtubule occupancy is converted to an inhibitory signal requires identifying the proteins in the kinetochore that modulate the checkpoint. The major conclusion from this work is that the conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and the spindle checkpoint. The state of kinetochore assembly is a caveat in interpreting all spindle checkpoint experiments, and it is important to have some measure of kinetochore integrity. Depletion of the Nuf2 homolog of C. elegans (HIM-10) using RNAi disrupts kinetochore structure by electron microscopy and causes a failure of bipolar spindle attachment (Howe et al. 2001). We found a similar dramatic effect on kinetochore structure when chromosomes were assembled in extracts depleted of xNdc80 and xNuf2. The RNAi experiments were performed by injecting dsRNA into the developing oocytes of hermaphrodites and observing the effects on subsequent embryonic cells. This experiment is similar to our antibody depletion experiments using Xenopus extracts because the kinetochores are assembled in the absence of xNuf2. In both cases there was an effect on kinetochore assembly. In contrast, adding function-blocking antibodies to extracts after kinetochores were assembled did not displace inner and outer kinetochore proteins. This demonstrates that adding antibodies to preassembled kinetochores did not affect associations of proteins to the kinetochore dramatically.
We have shown in multiple ways that the Ndc80 complex is required for spindle checkpoint signaling. Injecting either anti-xNdc80 or anti-xNuf2 antibodies into cultured cells affected chromosome congression and abrogated the spindle checkpoint. Addition of anti-xNdc80 antibody to Xenopus extracts with preassembled kinetochores also blocked checkpoint signaling without affecting many protein associations with the outer kinetochore. xRod and xZw10 are still associated with the kinetochore in these extracts, indicating that kinetochores can bind at least two spindle checkpoint proteins. Unfortunately, we could not check for the presence of the other spindle checkpoint proteins because of the identical source (rabbit). Martin-Lluesma et al. (2002) recently showed that inactivating HEC1 (Ndc80) by RNAi results in a checkpoint-dependent mitotic arrest, which seemingly contradicts our Xenopus results but is consistent with yeast single mutants lacking either Ndc80 or Nuf2. A model to reconcile all experiments is that antibody addition results in the inactivation of both xNdc80 and xNuf2, because we have shown that inactivation of both proteins in yeast abrogates the spindle checkpoint. This also implies that the function of the Nuf2 homolog (MPP1) was unaffected by HEC1 (Ndc80) RNAi. Our experiments demonstrate the importance of antibody inactivation experiments to complement other approaches such as immunodepletion and RNAi. Function-blocking antibodies have allowed us to dissociate the effects of Ndc80 in the spindle checkpoint and chromosome congression from the role played in kinetochore assembly.
The protein that docks the Mad1 and Mad2 proteins at the kinetochore has not been identified. Spc25 in yeast shows a two-hybrid interaction with the spindle checkpoint protein Mad1 (Newman et al. 2000), and Hec1 (Ndc80) has recently been shown to have a similar two-hybrid interaction with human Mad1 (Martin-Lluesma et al. 2002). RNAi of Hec1 also removes Mad1 from the kinetochore. We and others have been unsuccessful in demonstrating a biochemical interaction between the Ndc80 complex and a checkpoint protein (Martin-Lluesma et al. 2002). This is not surprising, as the interaction of Mad2 at the kinetochore has a half-life of 24 sec.
Genetic analysis in yeast identifies two different protein complexes as potential checkpoint regulators, CBF3 and Ndc80 (Gardner et al. 2001; Janke et al. 2001). CBF3 mutants dramatically affect kinetochore assembly, and therefore checkpoint effects are probably indirect (Cheeseman et al. 2001; He et al. 2001; Janke et al. 2001; Li et al. 2002). Mutations that eliminate Spc24 or Spc25 and double mutants lacking both Ndc80 and Nuf2 are checkpoint-defective (Janke et al. 2001; Wigge and Kilmartin 2001). However, mutants of the Ndc80 complex are different from CBF3 mutants in that they have a less dramatic effect on kinetochore assembly (He et al. 2001; Janke et al. 2001). Chromatin immunoprecipitation (ChIP) experiments using spc24-2 and ndc80-1 mutants show that Ndc10, Okp1, Mcm21, Ctf13, and Ctf19 are present at kinetochores, suggesting that the kinetochore is largely intact (He et al. 2001; Janke et al. 2001). However, assembly of some proteins does require the Ndc80 complex because proteins of the DUO/DAM/DASH complex (DDD complex) and Stu2 are missing (He et al. 2001; Janke et al. 2001, 2002). The lack of both microtubule binding and the spindle checkpoint in mutants deficient for the Ndc80 complex cannot be explained by the lack of either the DDD complex or Stu2 because chromosomes in mutants lacking these proteins bind microtubules and are checkpoint-proficient (Cheeseman et al. 2001; Severin et al. 2001).
There are numerous observations that suggest a conserved role for the Ndc80 complex in modulating kinetochore–microtubule interactions. In yeast, mutation of proteins in the Ndc80 complex cause severe nondisjunction of chromosomes, and real-time analysis of GFP-tagged chromosomes shows random centromere movements during metaphase (He et al. 2001). The simplest interpretation is that chromosomes are detached from the spindle. A lack of kinetochore–microtubule interaction could also explain the failure of chromosomes to congress to the metaphase plate in antibody-injected XTC cells. xNdc80 and xNuf2 localize at kinetochores from prometaphase through anaphase. Therefore, they are present on the kinetochore during all mitotic phases necessary for mediating microtubule attachments. This pattern is different from the localization of most spindle checkpoint proteins that are only present in earlier stages of mitosis (Choo 1997; Shah and Cleveland 2000). These highly conserved proteins are ideally suited to mediate the interface between the essential kinetochore function of binding microtubules and regulating the cell cycle in response to aberrant kinetochore function.
The xNdc80 complex is 190 kD, which leaves ∼60 kD of protein(s) unidentified. Although there are no obvious homologs of Spc24 and Spc25 in metazoans, it is possible that other proteins have assumed their functions. We predicted, based on the size of the xNdc80 complex, that adding back recombinant xNdc80 and xNuf2 would be insufficient to rescue the checkpoint defects in depleted extracts. As predicted, we could not rescue the depleted extracts with recombinant proteins expressed in either bacteria or in baculovirus (data not shown). Although the recombinant proteins may not have been properly folded, a more likely explanation is that we also depleted additional interacting proteins from the extract. We are presently isolating the xNdc80 complex from Xenopus extracts to identify additional interacting proteins responsible for complete complex function.
Materials and methods
Isolation of Xenopus Ndc80, Nuf2, Rod, and Zw10 clones
All chemicals were purchased from Sigma-Aldrich unless stated otherwise. All DNA restriction enzymes were purchased from New England Biolabs. Using these ESTs, we designed the following PCR primers to obtain a cDNA from a Xenopus stage-11.5–14 cDNA library: xNuf2 (CCCCATATGGACAAACT GACCTTTCCG) and the T3 promoter primer (TAACCCT CACTAAAGGGA); xNdc80 (CCCCATATGAGGAGGAGCT CTGTCACC) and the T3 promoter primer; and the N terminus of xZw10 (GGGGGATCCCATATGCAGAGTGCAGAAGAC TTG) and CCCGGATCCAAGCTTACTTTACTGTGTTATG TATTTC. The C terminus of xRod was amplified from the EST dab69d10.y1 (accession no. BG233529) using primers GGGG GATCCCATATGTTTTGGGGAATTATATCTGCAG and GG GGGATCCAAGCTTTTATATATCTGAATGATC. xNdc80 and xNuf2 PCR products were cloned into the NdeI–NotI sites of pET28 (Novagen) to yield pET 28-xNuf2 and pET 28-xNdc80. xRod and xZw10 fragments were cloned into the NdeI–HindIII sites of pET 28 to yield pET 28-xRod and pET 28-xZw10. The xNdc80 (AY168013) and xNuf2 (AY168014) sequences were deposited into GenBank.
Protein purification, antibody generation, purification, and immunoblotting
Immunoblotting was performed as described (Gardner and Burke 2000; Bolton et al. 2002). 6His-xNdc80, xNuf2, xZw10, and xRod fusion proteins were expressed in the Escherichia coli strain BL21 (DE3 pLysS; Novagen), purified and injected into rabbits to produce polyclonal sera (Covance). Antibodies were affinity-purified on the corresponding immunizing protein coupled to Sepharose (Amersham Biosciences) as described (Harlow and Lane 1988) and dialyzed into phosphate-buffered saline (PBS). xRod and xZw10 affinity-purified antibodies recognize a single band by immunoblot at ∼250 kD and 80 kD in Xenopus egg extracts, respectively (characterization will be described elsewhere). Affinity-purified anti-xNdc80 antibodies were biotinylated using a FluoReporter Mini-Biotin-XX Protein Labeling Kit (Molecular Probes), and affinity-purified xRod and xZw10 antibodies were conjugated using the Alexa Fluor 568 Monoclonal Antibody Labeling Kit (Molecular Probes) as described by the manufacturer.
A chicken peptide antibody against the N terminus of an xCenp-A (identified using a BLAST search for Xenopus ESTs similar to human Cenp-A) was generated against the peptide MRPGSTPPSRRKSRPPRRVC and affinity-purified on this peptide coupled to SulfoLink Coupling Gel (PIERCE). xCenp-A affinity-purified antibody recognizes a single band by immunoblot at ∼17 kD in a Xenopus egg extract.
Immunofluorescence and microinjection of XTC cells, chromosomes assembled in Xenopus extracts, and yeast
Monolayers of Xenopus tissue culture cells (XTCs) were grown, treated with drugs, and microinjected as described (Kallio et al. 2002). Anti-Ndc80, anti-Nuf2, and control rabbit antibodies were microinjected at needle concentrations of 5, 8, and 8 mg/mL, respectively. Fluorescent and time-lapse images were captured as described (Bolton et al. 2002; Kallio et al. 2002).
xNdc80 and xNuf2 were immunolocalized in XTC cells as described (Bolton et al. 2002). To visualize single chromosomes, XTC cells were treated with nocodazole for 3 h, collected by mitotic shake-off, swollen in 15 mM KCl, and centrifuged onto coverslips prior to fixation. To observe all kinetochores from a single cell or sperm chromatin (Figs. 4, 5; Supplemental Fig. 1), three-dimensional data sets were collected at 0.3-μm steps and then compressed into a single plane. The images in Figures 1E, 4C,D, and 5B were processed identically to compare levels of kinetochore staining.
For kinetochore assembly assays, Xenopus extracts were diluted 20-fold in BRB80 (80 mM K-PIPES at pH 6.8, 1 mM MgCl2, 1 mM EGTA) plus 0.25% Triton X-100, 1% formaldehyde, and 20% glycerol for 5 min, centrifuged onto poly-L-lysine-coated coverslips through a 30% glycerol cushion containing BRB80 plus 0.25% Triton X-100, and postfixed in −20°C methanol for 3 min. Chromatin was stained with antibodies against xMad1, xMad2, xBub1, and xBub3 as previously described (Chen et al. 1998) and with antibodies to xNdc80 (3 μg/mL), xNuf2 (3.5 μg/mL), xCenp-A (4 μg/mL), XKCM1 (2 μg/mL), P150 Glued (0.5 μg/mL; Transduction Laboratories), xRod (4 μg/mL), and xZw10 (4 μg/ml), Cy3-conjugated anti-xRod (5 μg/mL), and Cy3-conjugated anti-xZw10 (5 μg/mL) and the appropriate secondary antibodies from Jackson Research. Yeast immunofluorescence to stain for tubulin and chromatin was performed as previously described (Adams and Pringle 1984).
Production and use of Xenopus egg extracts
Adult wild-type Xenopus laevis were purchased from Nasco. CSF-arrested egg extracts, demembranated sperm nuclei, and H1 kinase assays were prepared as described (Murray 1991). Extracts were prepared and used as described (Minshull et al. 1994; Stukenberg et al. 1997).
Immunoprecipitations, column chromatography, and sucrose gradients were performed as described (Bolton et al. 2002). xNdc80 and xNuf2 were depleted from 150 μL of CSF-extracts by two consecutive incubations with 30 μL of Affi-prep protein A beads (Bio-Rad) covalently coupled to 30 μg of anti-xNdc80 antibody.
Yeast strains and media
Cells were grown in YM-1 medium (Hartwell 1967) and synthetic complete medium lacking essential nutrients as required (Gardner et al. 2001). BAR1 strains were arrested in G1 with 10 μM α-factor. Degron alleles were constructed and integrated as described (Dohmen et al. 1994). Cells were synchronized and analyzed as described previously (Gardner et al. 2001) Strains are congenic with W303a background except for 3026-2-2, 3027-1-4, 3054-3-2, and 3046, which are W303/A364a. W303a is MATa ura3-1 leu2-2,112 his3-11,15 trp1-1 ade2-1 can1-100 GAL+. The relevant genotypes of the strains used are: 3026-2-2, MATa ndc80-1 bub2∷HIS3; 3027-1-4, MATa nuf2-61 bub2∷HIS3; 3046, MATa nuf-61 mad2∷KANMX6 PDS1:13myc; 3047, MATa nuf2-61 PDS1:13myc; 3048, MATa ndc80-1 PDS1:13myc; 3054-3-2, MATa ndc80-1 mad2∷KANMX6 PDS1:13myc; 3050, MATa nuf2 dt:URA3 PGAL10-UBR1:HIS3 PDS1:13myc; 3051, MATa nuf2dt:URA3 PGAL10-UBR1:HIS3 ndc80-1 PDS1:13myc; 3052, MATa PDS1:13myc; 3053, MATa ndc80dt:URA3 PGAL10-UBR1:HIS3 PDS1:13myc.
Yeast cell synchronization and protein extraction
Yeast cells were synchronized as described (Gardner et al. 2001). To extract protein, cells were suspended in 0.1 M NaOH and incubated at room temperature for 5 min. They were pelleted, resuspended in SDS-PAGE sample buffer, boiled for 5 min, and cleared by centrifugation. The concentration of protein in the supernatant was determined by the Bradford assay after 2500-fold dilution, and equal amounts of protein were run on SDS-PAGE gels.
Acknowledgments
We thank John Kilmartin for providing plasmids and strains; Rey-Huei Chen for Mad1, Mad2, Bub1, and Bub3 antibodies; F. Soloman for the anti-Tub2; C. Walzcak for providing XKCM1 antibody; and the members of the Burke, Gorbsky, and Stukenberg labs for helpful comments and support. D.J.B., J.R.D., and R.D.G are supported by grants from the National Institutes of Health (GM40334) and the University of Virginia Cancer Center Support Grant (P30 CA44579). M.J.K. and G.J.G. are supported by a grant from the National Institutes of Health (GM50412). M.L.M. and P.T.S. are supported by the National Institutes of Health (GM63045) and the Pew Charitable Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pts7h@virginia.edu; FAX (434) 924-5069.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1040903.
Supplementary material is available at http://www.genesdev.org.
References
- Adams AE, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Gomes R, Karess R E. Rough Deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat Cell Biol. 2000;2:939–943. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- Bolton MA, Lan W, Powers SE, McCleland ML, Kuang J, Stukenberg PT. Aurora B kinase exists in a complex with Survivin and INCENP and its kinase activity is stimulated by Survivin binding and phosphorylation. Mol Biol Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MT, Goetsch L, Hartwell LH. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GKT, Jablonski SA, Starr DA, Goldberg ML, Yen TJ. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: Linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol. 2002;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo A. The centromere. New York: Oxford University Press; 1997. [Google Scholar]
- Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: Movement from inner centromere to midbody during mitosis. J Cell Biol. 1987;105:2053–2067. doi: 10.1083/jcb.105.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Gardner RD, Burke DJ. The spindle checkpoint: Two transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Gardner RD, Poddar A, Yellman C, Tavormina PA, Monteagudo MC, Burke DJ. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics. 2001;157:1493–1502. doi: 10.1093/genetics/157.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore–microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Howe M, McDonald KL, Albertson DG, Meyer BJ. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J Cell Biol. 2001;153:1227–1238. doi: 10.1083/jcb.153.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1–Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Bachant JB, Castillo AR, Giddings TH, Jr, Winey M. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol Biol Cell. 1999;10:2377–2391. doi: 10.1091/mbc.10.7.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes & Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA. Role of hec1 in spindle checkpoint signaling and kinetochore recruitment of mad1/mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: A link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- Newman JR, Wolf E, Kim PS. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc Natl Acad Sci. 2000;97:13203–13208. doi: 10.1073/pnas.97.24.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne MA, Schlenstedt G, Jinks T, Silver PA. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Saitoh H, Tomkiel J, Cooke CA, Ratrie H, III, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Severin F, Habermann B, Huffaker T, Hyman T. Stu2 promotes mitotic spindle elongation in anaphase. J Cell Biol. 2001;153:435–442. doi: 10.1083/jcb.153.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Cleveland DW. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Lustig KD, McGarry TJ, King RW, Kuang J, Kirschner MW. Systematic identification of mitotic phosphoproteins. Curr Biol. 1997;7:338–348. doi: 10.1016/s0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu F, Elledge SJ. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: Novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Steuer ER, Sheetz MP, Mitchison T. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J Cell Biol. 1991;114:285–294. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]