Abstract
Cells have evolved checkpoint responses to arrest or delay the cell cycle, activate DNA repair networks, or induce apoptosis after genomic perturbation. Cells have also evolved the translesion synthesis processes to tolerate genomic lesions by either error-free or error-prone repair. Here, we show that after a replication perturbation, cells exhibit a mutator phenotype, which can be significantly affected by mutations in the checkpoint elements Cds1 and Rad17 or translesion synthesis polymerases DinB and Polζ. Cells respond to genomic perturbation by up-regulation of DinB in a checkpoint activation-dependent manner. Moreover, association of DinB with chromatin is dependent on functional Rad17, and DinB physically interacts with the checkpoint-clamp components Hus1 and Rad1. Thus, translesion synthesis is a part of the checkpoint response.
Keywords: Checkpoint response, mutator phenotype, mutagenic translesion synthesis, DinB, Cds1, Rad17
Organisms, when faced with genomic stress, activate checkpoints to regulate cell-cycle transitions and facilitate DNA repair processes, or to induce apoptosis (Elledge 1996; Zhou and Elledge 2000). Failure in these responses can result in the accumulation of mutations and cause genomic instability, which are predominant characteristics of cancer cells (Hartwell and Kastan 1994; Lengauer et al. 1998). To achieve the checkpoint response, cells have sensors/transducers that detect genomic lesions and relay the signals to effector kinases that act on the targets (Zhou and Elledge 2000). In fission yeast, Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1, known as the checkpoint Rads, function as sensors/transducers for signals of replication perturbation and DNA damage (Rhind and Russell 1998; Caspari and Carr 1999). Rad3 plays a central role in the checkpoint signaling pathway, forming a complex with Rad26 in vivo, and phosphorylating Rad26 in response to DNA damage and late S-phase arrest (Edwards et al. 1999; O'Connell et al. 2000). Rad1, Rad9, and Hus1 exist in a complex in vivo and have predicted structural homology to PCNA (Caspari et al. 2000; Venclovas and Thelen 2000; Melo and Toczyski 2002). Rad17 contains homology to all five subunits of RF-C (Griffiths et al. 1995) and exists in a complex with Rfc2-5 (for review, see Caspari and Carr 1999; Melo and Toczyski 2002). A fraction of fission yeast Rad17 binds to chromatin throughout the cell cycle, independent of the presence of other checkpoint proteins (Griffiths et al. 2000). Increasing amounts of Rad17 bind to chromatin in response to DNA damage and remain tightly chromatin bound after replication arrest (Kai et al. 2001). In budding yeast, Ddc1 (fission yeast Rad9 homolog) recognizes damage sites in a Rad24 (fission yeast Rad17 homolog)-dependent manner (Kondo et al. 2001; Melo et al. 2001), whereas inhibition of human Rad17 synthesis by siRNA leads to a reduction in the damage-induced binding of human Rad9 to chromatin (Zou et al. 2002). These data have led to a hypothesis that the Rad17–Rfc2-5 complex senses genomic lesions and functions as a checkpoint-clamp loader, loading the checkpoint-clamp Rad1–Rad9–Hus1 complex onto chromatin. Once loaded onto chromatin, the checkpoint-clamp may recruit other checkpoint elements to activate the checkpoint response (Melo and Toczyski 2002). In addition to the checkpoint Rad proteins, the fission yeast checkpoint effector kinase Cds1 and its budding yeast counterpart Rad53 play a critical role in preventing accumulation of abnormal replication intermediates when replication is perturbed in order to restore the replication fork and to allow restart of DNA synthesis (Desany et al. 1998; Lindsay et al. 1998; Rhind and Russell 2000; Lopes et al. 2001). Thus, checkpoint rad+ genes and cds1+ are essential in coordinating DNA replication and maintaining genomic stability.
Cells have also evolved a process that allows them to tolerate genomic lesions by either error-free or error-prone translesion synthesis (TLS). In response to genotoxic stress, Escherichia coli has an SOS response that up-regulates genes to cope with the stress (Friedberg et al. 1995; Sutton et al. 2000; Goodman 2002). Many of the gene products induced by the SOS response are involved in DNA repair. Of these, three SOS genes—umuC, umuD, and dinB—are exclusively responsible for the TLS process (Sutton et al. 2000; Goodman 2002). The UmuC protein associates with a proteolytically processed form of UmuD (UmuD‘) and forms a UmuD‘2C complex (PolV). In vitro, the UmuD‘2C complex bypasses certain lesions in a highly error-prone manner and has poor fidelity in copying undamaged DNA (Sutton et al. 2000; Goodman 2002). The gene product of dinB (PolIV) is a DNA polymerase that is devoid of 3′–5′ exonuclease activity, distributive in nature, and prone to frameshift and that frequently introduces −1 frameshift mutations (Wagner et al. 1999; Goodman 2002).
A large repertoire of eukaryotic genes known or predicted to encode translesional DNA polymerases has emerged in recent years (Woodgate 1999; Friedberg et al. 2000; Ohmori et al. 2001; Goodman 2002). Among these polymerases, Polζ is a B-family polymerase required for DNA damage-induced mutagenesis in budding yeast (Goodman 2002), which has the ability to bypass a cys–syn thymine–thymine (T–T) dimer (Nelson et al. 1996). Some of the other translesional polymerases are members of the Y-family or UmuC-super family, represented by UmuC, Rev1, DinB/PolIV/Polκ, Rad30/Polη/Eso1, and Polι (Ohmori et al. 2001; Wang 2001; Goodman 2002). Rad30/Polη performs translesion DNA synthesis of cys–syn T–T dimers with high fidelity. However, Rad30/Polη exhibits extremely low fidelity when acting on undamaged DNA or on DNA with noncognate substrates (Johnson et al. 1999; Masutani et al. 1999; Johnson et al. 2000; Matsuda et al. 2001; Washington et al. 2001; Goodman 2002). In fission yeast, the eso1+ gene encodes a dual function protein, with the N-terminal portion encoding the homolog of Rad30/Polη and the C-terminal region encoding the budding yeast Eco1 homolog (Tanaka et al. 2000). In vitro, human DinB/Polκ has a high misincorporation rate and can bypass abasic sites and N-2-acetylaminofluorene (AAF) adducts in a highly error-prone manner (Ohashi et al. 2000a,b). Polκ/DinB can also readily extend a misaligned undamaged primer terminus, causing −1 frameshift mutations (Washington et al. 2002). However, it bypasses thymine glycols with a preference for correct nucleotides incorporation (Fischhaber et al. 2002). Given the error-prone nature of these translesion synthesis polymerases in copying undamaged DNA and noncognate damaged-substrates (Friedberg et al. 2002; Goodman 2002), it seems likely these polymerases are a source of mutagenesis. Thus, understanding the regulation of the action of translesional polymerases will help to clarify how cells maintain normal genomic stability.
Studies of the E. coli SOS response have suggested that the umuCD gene products play two temporally separate physiological roles in the DNA damage checkpoint and TLS (Opperman et al. 1999). Little is known in eukaryotes about how TLS is regulated and its relationship with the checkpoint response. Genetic studies in budding yeast have shown that UV-induced mutagenic repair of irreparable lesions requires checkpoint function (Paulovich et al. 1998), whereas the activation of budding yeast S-phase checkpoint genes DUN1 or RFC5 correlates with an increase of mutation rate in the pol3-01 mutant (Datta et al. 2000). These studies suggest that when cells encounter an irreparable lesion, the checkpoint response may activate translesion synthesis to tolerate the lesion by mutagenic repair.
We have previously shown that a large repertoire of thermo-sensitive fission yeast replication mutants exhibits a mutator phenotype at the semipermissive temperature. The types of mutations are characterized by deletion of sequences flanked by homologous repeats and small sequence alterations comprised of base substitutions and single-base frame shifts (Liu et al. 1999). Replication perturbation caused by a semidisabled replication mutant could generate a single-strand gap, leading to a double-strand break. Subsequent repair by double-strand break repair, recombinational repair, or single-strand annealing could result in deletion of sequences flanked by homologous repeats (Paques and Haber 1999). In addition, a stalled replication fork caused by an impaired replication mutant could induce regression of the replication fork and formation of a Holliday junction (Paques and Haber 1999; Boddy et al. 2001). Resolution of the Holliday junctions could also cause a deletion mutator phenotype.
Small sequence alterations are often caused by polymerase slippage or misinsertions by mutations in DNA polymerases. Here, we use a fission yeast polα thermo-sensitive mutant, swi7-H4, to induce replication stress and a mutator phenotype at the semipermissive temperature. By analyzing the effect of the checkpoint proteins, Cds1 and Rad17, on the mutation rate and mutation spectra in the polα(swi7-H4) mutant, we find that Cds1 and Rad17 significantly affect its mutator phenotype. A mutation in cds1+ enhances the deletion mutations in the polα(swi7-H4) mutant, whereas mutations in rad17+ have a suppressive effect on both deletion and point mutations. Analysis of the deletion of three translesional polymerases in polα(swi7-H4) revealed that DinB, and Polζ to a lesser extent, contribute significantly to the induction of point mutations and single-base frame shifts. Further studies showed that activation of the checkpoint plays a significant role in regulating the translesional polymerase, DinB.
Results
Checkpoint effector kinase Cds1 affects the mutator phenotype in polα (swi7-H4).
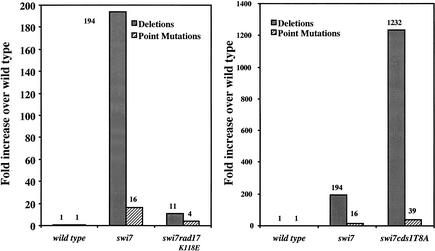
We used a replicative polymerase mutant polα(swi7-H4) to induce replication stress and a mutator phenotype. This mutant exhibited an S-phase delay and activation of Cds1 kinase at its semipermissive temperature (30°C), indicating that the checkpoint is intact in polα(swi7-H4) (Fig. 1A,B). polα(swi7-H4) exhibited an elevated mutation rate compared with wild-type cells at the semipermissive temperature (Table 1). The mutant accumulated both deletions of sequence flanked by repeats and small sequence alterations such as base substitutions and frame shifts (hereafter, all small sequence alterations are referred to as point mutations) and exhibited a 194-fold increase in deletion mutations and 16-fold increase in point mutations when compared with wild-type cells (Fig. 2).
Figure 1.
Analysis of checkpoint function in polα(swi7-H4), polα(swi7-H4) rad17.K118E, and polα(swi7-H4) cds1.T8A cells. (A) Progression of wild-type, polα(swi7-H4), polα(swi7-H4) rad17.K118E, and polα(swi7-H4) cds1.T8A cells through S phase. Wild-type and mutant cells were synchronized by nitrogen starvation for 24 h in G1 phase and then released into synthetic media containing nitrogen. The X-axis represents the relative DNA content, and the Y-axis represents cell number at the indicated time after release from nitrogen starvation. (B) Activation of Cds1 kinase in checkpoint mutants. Logarithmically growing polα(swi7-H4) and polα(swi7-H4) mutant cells in checkpoint mutant backgrounds were incubated at 30°C for 4 h, and the Cds1 kinase activity was assayed from the cell extracts as described in Materials and Methods. Levels of Cds1 used in the kinase assay were determined by the Cds1 immunoblot. The polα(swi7-H4) mutant is presented as swi7.
Table 1.
Effects of Cds1 and Rad17 on the polα(swi7-H4) mutation rate
| Genotype
|
Mutation rate (×10−8)
|
C.I.
|
Fold increase over wild type
|
Mutation rate relative to swi7
|
|---|---|---|---|---|
| wild type | 5.70 | (4.98–6.12) | 1 | |
| Wild type +Cds1 | 3.98 | (3.61–5.22) | 1 | |
| swi7 | 198 | (155–233) | 35 | |
| swi7 +Cds1 | 10.3 | (7.59–12.5) | 2 | ↓19 |
| cds1.T8A | 6.22 | (4.55–6.71) | 1 | |
| swi7 cds1.T8A | 911 | (796–1305) | 160 | ↑4.6 |
| swi7 rad17.K118E | 25.9 | (20.3–30.1) | 5 | ↓7.6 |
| swi7 rad17.E198A | 83.5 | (64.7–103) | 15 | ↓2.4 |
| swi7 rad17.S222A | 68.2 | (59.2–72.5) | 12 | ↓2.9 |
| rad17.K118E | 5.43 | (4.68–6.33) | 1 | |
| rad17.E198A | 5.72 | (4.42–6.59) | 1 | |
| rad17.S222A | 5.21 | (4.41–6.29) | 1 |
Mutation rates shown here are the average of three or more experiments. C.I. is the 95% confidence interval (10−8 per cell division). Fold increases or decreases in mutation rates are shown as ↑ or ↓, respectively. +Cds1 represents ectopic expression of Cds1 from pREP4l-cds1+.
Figure 2.
Types of mutations in polα(swi7-H4) cells in rad17.K118E and in cds1.T8A backgrounds. Point mutations represent both base substitutions and single-base frameshift mutations. The rates of each mutation type were calculated by multiplying the rate of 5-FOA-resistant cells by the percentage of alterations that occurred as deletion or point mutations. Bar graphs show the types of mutation relative to wild-type. The polα(swi7-H4) mutant is presented as swi7.
Cds1, the checkpoint effector kinase, is thought to play a critical role in recovery from a stalled replication fork to restart DNA synthesis (Desany et al. 1998; Lindsay et al. 1998; Rhind and Russell 2000; Lopes et al. 2001; Tercero and Diffley 2001). Ectopic expression of Cds1 effectively suppresses the mutation rate in polα(swi7-H4) to near wild-type level (Table 1), consistent with the notion that Cds1 somehow prevents accumulation of aberrant replication intermediates and promotes recovery of the replication fork.
Because deletion of cds1+ in the polα mutant cells is lethal (Bhaumik and Wang 1998), we tested the effect of a cds1 mutant, cds1.T8A, which retains Cds1 kinase activity and is only moderately sensitive to hydroxyurea (HU) treatment (Tanaka et al. 2001). The double mutant polα(swi7-H4) cds1.T8A had a moderately compromised checkpoint, completing the cell cycle after 7 h (Fig. 1A) and had greater than 80% viability at the semipermissive temperature (30°C). In addition, Cds1 kinase activity was weakly induced in the polα(swi7-H4) cds1.T8A double mutant (Fig. 1B). However, polα(swi7-H4) cds1.T8A exhibited an overtly elevated mutation rate that was 160-fold higher than wild-type cells and 4.6-fold higher than the polα(swi7-H4) single mutant (Table 1). Moreover, the polα(swi7-H4) cds1.T8A double mutant accumulated a sixfold higher deletion mutations and a greater than twofold higher point mutations than the polα(swi7-H4) single mutant (Fig. 2, right). These results support the notion that Cds1 prevents accumulation of abnormal replication intermediates that could lead to deletion mutations.
To confirm this notion, we analyzed another checkpoint rad mutant, rad26.T12, which has been shown to play a role in recovery from DNA damage (Al-Khodairy et al. 1994; Lindsay et al. 1998). Similar to the effect of the cds1.T8A mutation, the mutation rate in the double mutant polα(swi7-H4) rad26.T12, was 147-fold higher than wild-type and 4.2-fold higher than the polα(swi7-H4) single mutant (data not shown). These results indicate that checkpoint gene products involved in replication fork maintenance and recovery are important in preventing genomic instability in coordination with DNA replication.
Mutations in rad17+ and cds1+ have opposite effects on the polα (swi7-H4) mutator phenotype
Studies of fission yeast and human cells have shown that genomic perturbation enhances chromatin binding of Rad17, which is thought to initiate the checkpoint response by promoting the binding of the checkpoint clamp to chromatin (Kai et al. 2001; Zou et al. 2002). We thus analyzed the effect of Rad17 on the mutator phenotype in polα(swi7-H4). Because deletion of rad17+ in polα mutants is lethal (Bhaumik and Wang 1998), we analyzed the effect of three rad17 mutants: rad17.K118E, rad17.E198A, and rad17.S222A. These rad17 mutants contain mutations in the Walker domains of the Rad17 protein and exhibit a moderate checkpoint defect (Griffiths et al. 1995). In contrast to the effect of the cds1.T8A mutation, mutations in rad17+ at all three alleles in polα(swi7-H4) suppressed the mutation rate compared with the polα(swi7-H4) single mutant, with rad17.K118E exhibiting the most significant effect (Table 1). Both deletion and point mutations in polα(swi7-H4) were suppressed by the rad17.K118E mutation to near wild-type level (Fig. 2, left). The suppression is not owing to a further compromise of the checkpoint function of the double mutant, because at the semipermissive temperature, the double mutant polα(swi7-H4) rad17.K118E exhibited a delay in S phase similar to that of the polα(swi7-H4) single mutant (Fig. 1A), and an ability to activate Cds1 kinase activity (Fig. 1B). However, Cds1 was not autophosphorylated in the polα(swi7-H4) rad17.K118E double mutant (Fig. 1B), indicating that the rad17.K118E mutation in polα(swi7-H4) compromised some physiological function other than checkpoint signaling for cell-cycle arrest. Although the double mutant, polα(swi7-H4) rad17.K118E, was able to maintain the checkpoint at 30°C, it exhibited a cut phenotype at 36°C, indicating lack of a functional checkpoint at the elevated temperature.
These experiments indicate that Rad17 and Cds1 act in opposite ways in coping with replication stress.
Translesional polymerases DinB and Polζ are responsible in generating the point mutations accumulated in polα (swi7-H4) cells
The ability of mutations in rad17+ to suppress the mutator phenotype of polα(swi7-H4) implies that Rad17 must play a positive role in enhancing mutations in polα(swi7-H4) when replication is perturbed. One possibility is that Rad17 promotes translesion synthesis by using the specialized translesional DNA polymerases for mutagenic synthesis. To test this hypothesis, we constructed mutants containing a deletion of polζ+, the rad30+ domain (Polη domain) of eso1+, or dinB+ individually and in combination with polα(swi7-H4) (Table 2). Deletion of polζ+ in polα(swi7-H4) moderately reduced the mutation rate to a level 25-fold over wild-type cells compared with 35-fold in the polα((wi7-H4) single mutant. Deletion of the rad30+ domain of eso1+ in polα(swi7-H4) did not significantly change the mutation rate. Deletion of dinB+ in polα(swi7-H4) reduced the mutation rate to 1.6-fold lower than that of the polα(swi7-H4) single mutant (Table 2). The triple mutant of polα(swi7-H4) with deletions of both polζ+ and dinB+ [polα(swi7-H4) polζΔ dinBΔ] exhibited a further decrease in mutation rate to a level twofold lower than the mutation rate of polα(swi7-H4) single mutant. The triple mutants polα(swi7-H4) rad30Δ dinBΔ and polα(swi7-H4) rad30Δ polζΔ exhibited a mutation rate similar to that of the double mutants polα(swi7-H4) dinBΔ and polα(swi7-H4) polζΔ, respectively (Table 2). These results indicate that both DinB and Polζ contribute to the elevated mutation rate in polα(swi7-H4) at 30°C, whereas the rad30+ domain of eso1+ does not contribute to the elevated mutation rate in polα(swi7-H4) when replication is under stress.
Table 2.
Deletion of dinB+reduces the mutation rate
| Genotype
|
Mutation rate (×10−8)
|
C.I.
|
Fold increase over wild type
|
Mutation rate relative to swi7
|
|---|---|---|---|---|
| wild type | 5.70 | (4.98–6.12) | 1 | |
| swi7 | 198 | (155–233) | 35 | 1 |
| swi7 polζΔ | 140 | (121–165) | 25 | ↓1.4 |
| swi7 rad30Δ | 175 | (161–231) | 31 | |
| swi7 dinBΔ | 124 | (109–162) | 22 | ↓1.6 |
| swi7 polζ dinBΔ | 91.8 | (72.3–122) | 16 | ↓2.0 |
| swi7 polζΔrad30Δ | 151 | (138–180) | 27 | ↓1.3 |
| swi7 rad30ΔdinBΔ | 130 | (106–171) | 23 | ↓1.5 |
| polζΔ | 5.01 | (4.20–5.91) | 1 | |
| rad30Δ | 13.4 | (10.2–16.1) | 2.3 | |
| dinBΔ | 4.18 | (3.32–5.22) | <1 | |
| polζΔ dinBΔ | 4.01 | (3.33–4.99) | <1 | |
| polζΔ rad30Δ | 11.5 | (9.9–13.6) | 2.0 | |
| rad30Δ dinBΔ | 10.2 | (8.9–14.2) | 1.8 |
Mutation rates are the average of three or more experiments. C.I. is the 95% confidence interval (10−8 per cell division). Fold decreases in mutation rates are shown as ↓.
Analysis of the mutation spectra of these double and triple mutants further confirmed the contribution of DinB and Polζ to the mutator phenotype seen in polα(swi7-H4) (Fig. 3). Deletion of dinB+ in polα(swi7-H4) reduced the level of point mutation from 16.2-fold to 4.8-fold over that of the wild-type. In contrast, deletion of either dinB+ or polζ+ in polα(swi7-H4) did not affect the levels of deletion mutations in cells (data not shown). The double-mutant polα(swi7-H4) dinBΔ had a comparable level of point mutations to that observed in polα(swi7-H4) rad17.K118E, which suppresses the point mutations (Fig. 3). The triple mutant polα(swi7-H4) dinBΔ polζΔ exhibited a further reduction of point mutation levels to near wild-type level (Fig. 3). The polα(swi7-H4) rad30Δ double mutant had a similar point mutation level as the polα(swi7-H4) single mutant. The triple mutant polα(swi7-H4) rad30Δ polζΔ had similar levels of point mutations as polα(swi7-H4) polζΔ, whereas the polα(swi7-H4) dinBΔ rad30Δ triple mutant had similar levels of point mutations as the polα(swi7-H4) dinBΔ double mutant (Fig. 3).
Figure 3.
Point mutation rates of polα(swi7-H4) mutant with deletions of translesional polymerases. The point mutation rate was calculated by multiplying the rate of 5-FOA-resistant cells by the percentage of point mutations that occurred. Bar graphs show the mutation rate of each mutant relative to wild-type cells. The polα(swi7-H4) mutant is presented as swi7.
Together, these results indicate that DinB and Polζ are responsible in generating the elevated mutation rate by accumulating point mutations when replication is perturbed in polα(swi7-H4), with DinB contributing the most to the mutator phenotype.
Up-regulation of DinB depends on checkpoint-activation in response to genomic perturbation
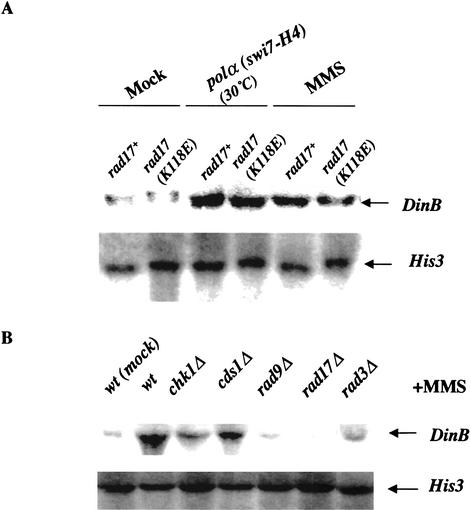
The rad17.K118E mutation in polα(swi7-H4) suppressed the mutator phenotype to a level similar to that of deletion of dinB+ in polα(swi7-H4) under replication stress (cf. Tables 1 and 2; Fig. 3). It is possible that the rad17.K118E mutation has a suppressive effect on the DinB function, resulting in a phenotype similar to that of dinB+ deletion. These have prompted us to analyze how DinB is regulated in cells after genomic perturbation. The level of dinB+ transcripts from cells under replication stress and from cells treated with methyl methansulfonate (MMS) were compared to unperturbed wild-type cells by Northern analysis. Transcript levels of dinB+ were also compared with and without replication stress or DNA damage in the rad17.K118E mutant. The dinB+-transcript (∼1.7 kb) was up-regulated in response to replication stress and DNA damage. The rad17.K118E mutation did not affect the up-regulation of dinB+ transcript in response to either replication stress or DNA damage (Fig. 4A). Because the checkpoint response is somewhat intact and Cds1 can be activated by HU in the rad17.K118E mutant (Kai et al. 2001), the up-regulation of dinB+ transcript may be enforced by the residual checkpoint function in the rad17.K118E mutant. To test this possibility, we analyzed the dinB+ transcript expression in cells deleted for chk1+, cds1+, rad9+, rad17+, and rad3+ after MMS treatment (Fig. 4B). The dinB+ transcript was up-regulated in wild-type cells after MMS treatment and is moderately up-regulated in cells with cds1Δ. In contrast, MMS treatment failed to induce the expression of dinB+ transcript in cells with a deletion of chk1+, rad3+, rad9+, or rad17+ (Fig. 4B).
Figure 4.
Expression of DinB is dependent on checkpoint activation. (A) DinB transcription is up-regulated in response to damage and replication perturbation. Logarithmically growing wild-type and rad17.K118E cells were incubated in 0.025% MMS for 4 h. Replication perturbation was induced in polα(swi7-H4) and polα(swi7-H4) rad17.K118E cells by culturing for 4 h at 30°C. Forty micrograms of total RNA was prepared from each strain and analyzed by Northern blotting. (B) Up-regulation of DinB transcript is dependent on checkpoint activation. Wild-type and checkpoint mutants cells were MMS treated for 2 h as in A. Forty micrograms of total RNA was used for Northern analysis. His3+ was used as a loading control. (C) DinB protein level is up-regulated in response to damage and replication perturbation. Logarithmically growing wild-type:dinB+-myc and rad17Δ:dinB+-myc cells were MMS treated for 2 h as described in A. Replication in polα(swi7-H4):dinB+-myc and polα(swi7-H4) rad17.K118E:dinB-myc cells was perturbed at 30°C as described in A. Cell extracts were prepared from the MMS-treated or replication-perturbed cells under denaturing conditions. DinB-myc was visualized by immunoblotting 40 μg of total cell extract protein with anti-myc antibodies. (D) Up-regulation of DinB protein is dependent on the checkpoint activation. Strains with dinB+-myc in checkpoint deletion mutant backgrounds were MMS-treated for 2 h as above. Forty micrograms of cell extract protein from wild-type cells and mutant cells was analyzed by immunoblotting using the anti-myc antibody with α-tubulin as a loading control.
DinB protein levels from wild-type cells or polα(swi7-H4) mutant with or without the rad17.K118E mutation were analyzed with a chromosomal integrated myc-tagged dinB+. Similar to the induction of dinB+-transcript expression, DinB protein levels were up-regulated in response to replication perturbation induced by culturing polα(swi7-H4) at 30°C or by MMS treatment (Fig. 4C). To test whether up-regulation of DinB protein is dependent on checkpoint activation, we constructed cds1Δ, chk1Δ, rad9Δ, rad17Δ, and rad3Δ strains containing a chromosomal integrated dinB+-myc. The DinB protein level was up-regulated in wild-type cells and in cds1Δ cells after MMS treatment. However, in cells with a deletion of chk1+, rad9+, rad17+, or rad3+, the DinB protein level was not up-regulated after MMS treatment (Fig. 4D).
Together, these results indicate that both the transcript and the protein level of DinB are up-regulated in response to genomic perturbation and this up-regulation depends on checkpoint-activation.
Association of DinB with chromatin correlates to the Rad17 chromatin-binding ability
Although DinB protein is expressed in the rad17.K118E mutant in response to genomic perturbation (Fig. 4C), the rad17.K118E mutation in polα(swi7-H4) has a suppressive effect on the mutator phenotype (Table 1; Fig. 2). This suggests that the expressed DinB polymerase in the polα(swi7-H4) rad17.K118E double mutant was unable to perform mutagenic synthesis.
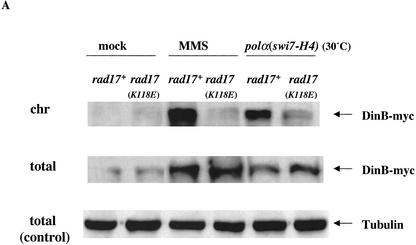
Our previous studies have shown that the rad17.K118E mutation compromised the ability of the Rad17 mutant protein to bind chromatin (Kai et al. 2001). This led us to speculate that the observed suppression of the polα mutator phenotype by the rad17.K118E mutation is owing to failure of DinB to associate with chromatin. To test this possibility, we analyzed chromatin association of DinB in response to replication perturbation and DNA damage in the rad17.K118E mutant background (Fig. 5A). DinB-myc associated with chromatin after MMS treatment or replication perturbation induced by culturing polα(swi7-H4) at 30°C. However, DinB chromatin association was severely compromised in the rad17.K118E mutant background (Fig. 5A).
Figure 5.
Chromatin association of DinB depends on the Rad17-chromatin-binding ability, and DinB interacts with checkpoint clamp components Hus1 and Rad1 in vivo. (A) Chromatin association of DinB in response to damage and replication perturbation depends on the Rad17-chromatin-binding ability. Logarithmically growing wild-type cells harboring dinB+-myc and rad17Δ:dinB+-myc cells were treated with MMS as described in Figure 4. polα(swi7-H4):dinB+-myc and polα(swi7-H4) rad17.K118E:dinB+-myc mutants were grown at 30°C to induce replication perturbation. Chromatin fractionation assay was performed as in Kai et al. (2001). Forty micrograms of total protein (total) and five volume equivalents of chromatin-bound protein (chr) were fractionated on SDS-PAGE, followed by immunoblotting. DinB-myc was visualized by using the anti-myc antibody. (B) DinB coprecipitates with Hus1. Crude cell lysates were prepared from 5 × 108 wild-type cells harboring hus1+-myc and dinB+-HA at their endogenous chromosomal loci. Cells were MMS treated as in Figure 4, and polα(swi7-H4):dinB+-HA:hus1+-myc and polα(swi7-H4) rad17.K118E:dinB+-HA:hus1+-myc cells were incubated for 4 h at 25°C or 30°C. (Bottom) Cell lysates prepared from wild-type cells harboring an untagged dinB+ and hus1+-myc or from cells with dinB+-HA and hus1+-myc treated with MMS were used as control. Cell lysates (300 μL) containing 1 mg protein were preincubated with 100 U of DNaseI and used for immunoprecipitation (IP) with the anti-HA antibody. The immunoprecipitates were immunoblotted with the anti-HA (left) or anti-myc monoclonal antibody (right). (C) Coprecipitation of DinB with Rad1 requires a functional Rad17. Crude cell lysates were prepared from 5 × 108 wild-type cells (rad17+) and rad17.K118E mutant cells harboring dinB+-myc and rad1+-HA at their endogenous chromosomal loci. Cells were treated with MMS as above, and the immunoprecipitation was performed as in B. The immunoprecipitates were immunoblotted with the anti-myc (left) or anti-HA monoclonal antibody (right). (Bottom) Control immunoprecipitation from lysates of MMS-treated wild-type cells harboring untagged dinB+ and rad1+-HA or rad1+-HA and dinB+-myc.
To test whether DinB chromatin binding requires a physical interaction with Rad17, a strain containing dinB+-myc and rad17+-HA at their respective genomic loci was constructed. DinB-myc failed to coimmunoprecipitate with the Rad17-HA protein after MMS treatment. This suggests that if there is any interaction between DinB and Rad17, it is transient or indirect.
DinB coprecipitates with checkpoint-clamp components Hus1 and Rad1
Failure of DinB to bind chromatin in the rad17.K118E mutant background after DNA damage or replication perturbation raises the question of whether Rad17 directly affects DinB chromatin association or whether the interaction is indirect through the checkpoint-clamp which is thought to be loaded onto chromatin by Rad17. We constructed a strain containing dinB+-HA and hus1+-myc at their respective genomic loci. Because fission yeast Hus1 exists in many forms (Caspari et al. 2000) and DinB is present in low abundance in cells, it is difficult to detect the DinB-HA in the anti-Hus1-myc-immunoprecipitate. However, Hus1-myc was readily detected in the anti-DinB-HA-immunoprecipitates from DNA-damaged cells and from replication perturbed cells, but not in undamaged or unperturbed cells (Fig. 5B, right panel). The coprecipitation of DinB-HA and Hus1-myc was not caused by association with DNA, because the coprecipitation experiments were carried out with DNase-treated cell lysates. Furthermore, the coprecipitation of DinB-HA and Hus1-myc was not owing to cross-reactivity of the antibody with the epitope tags. DinB-HA was not detected in the anti-DinB-HA-immunoprecipitates from lysates of cells containing an untagged dinB+ and hus1+-myc, whereas DinB-HA and Hus1-myc were both detected in the anti-HA-immunoprecipitates from lysates of cells containing dinB+-HA and hus1+-myc (Fig. 5B, lower panel).
To further verify the physical association of DinB and the checkpoint-clamp component in response to genomic perturbations, strains containing myc-tagged dinB+ and HA-tagged rad1+ at their respective genomic locus were constructed either in rad17+ or in the rad17.K118E mutant background. On MMS treatment, Rad1-HA was readily detectable in the anti-DinB-myc immunoprecipitates from rad17+ cells but not in the rad17.K118E mutant cells (Fig. 5C). Identical controls were performed as described above in DinB and Hus1 coprecipitation (Fig. 5C, lower panel). Because strain with an epitope tagged rad9+ exhibited a mild checkpoint defect, coprecipitation of DinB and Rad9 was not performed.
These results suggest that in response to replication stress or DNA damage, the association of DinB with chromatin for mutagenic translesion synthesis is mediated by its interaction with the checkpoint-clamp loaded by the checkpoint-clamp loader containing a functional Rad17. Thus, a prerequisite of regulating DinB expression and chromatin association for mutagenic translesion synthesis is checkpoint activation.
Discussion
In this study, we used a DNA polymerase α mutant, polα(swi7-H4), to perturb DNA replication at the semipermissive temperature (30°C). At the semipermissive temperature, polα(swi7-H4) exhibited a delay of cell-cycle transition, activation of Cds1 kinase, and Cds1 autophosphorylation, indicating that the checkpoint is intact in the mutant (Fig. 1A,B). Despite having an intact checkpoint, polα(swi7-H4) exhibits an elevated mutation rate and accumulates both deletion mutations and point mutations in cells. The mutator phenotype of polα(swi7-H4), however, is significantly affected by mutations in cds1+ and rad17+. Further analyses indicate that at 30°C, polα(swi7-H4) induces the expression of the translesional polymerase DinB in generating point mutations in a checkpoint activation dependent manner. Translesional polymerases are thought to function primarily in bypassing lesions on DNA template (Sutton et al. 2000; Goodman 2002). A replicative DNA polymerase is expected to generate misincorporations, single-stranded gaps on template, or mispaired primer termini, but not “lesions” on the template. Finding that a mutant of replicative polymerase induces the expression of DinB to generate point mutations is a novel mutation consequence found in replicative polymerase, in which an error-prone translesional polymerase is induced for mutagenic synthesis on a template without a lesion.
How might mutations in rad17+ affect point mutations in polα (swi7-H4)?
Here, we demonstrate that in response to genomic stress, up-regulation of both the transcript and protein levels of DinB (Fig. 4) is dependent on rad3+, rad9+, and rad17+ (Fig. 4). Because rad3+, rad9+, and rad17+ are the checkpoint sensors/transducers required for activation of the checkpoint, these data indicate that checkpoint activation is a prerequisite for DinB expression in response to replication stress or DNA damage.
After replication perturbation or DNA damage, DinB is expressed in the rad17.K118E mutant (Fig. 4). However, the rad17.K118E mutation notably suppresses the point mutations in polα(swi7-H4) cells (Table 1; Fig. 2). The lysine118 (Lys118) residue of Rad17 is localized in the Walker A domain of the Rad17 protein (Griffiths et al. 1995). Mutation of Lys118 to glutamate severely reduces the ability of the Rad17(K118E) mutant protein to form a complex with Rfc2 and to associate with chromatin (Kai et al. 2001). Furthermore, mutation at the equivalent lysine residue in human Rad17 also abolishes the ability of human Rad17 protein to associate with chromatin (Zou et al. 2002). After genomic perturbation, restart of a stalled replication fork through a mutagenic translesion synthesis process would require DinB to associate with chromatin. Despite being expressed in the rad17.K118E mutant, DinB protein is unable to associate with chromatin to perform the mutagenic translesion synthesis (Fig. 5A), as evident by the suppression of point mutations in polα(swi7-H4). This suggests that DinB chromatin association for mutagenic synthesis requires functional Rad17. Because deletion of any checkpoint-clamp component is lethal in the polα mutant (Bhaumik and Wang 1998) and checkpoint-clamp mutants are not yet available for this study, the effect of mutations in checkpoint-clamp on the mutator phenotype in polα(swi7-H4) is not yet known.
Comparison of the suppressive effect on mutation rate of the rad17.K118E mutation in polα(swi7-H4) with deletions of dinB+ and polζ+ in polα(swi7-H4) shows that mutation in rad17.K118E appears to be more suppressive than dinBΔ and polζΔ. It is important to mention that rad17.K118E mutation in polα(swi7-H4) suppresses both deletion and point mutations (Fig. 2, left panel), whereas deletions of both dinB+ and polζ+ in polα(swi7-H4) affect only the point mutation formation. Thus, the polα(swi7-H4) rad17.K118E double mutant exhibits a 7.6-fold decrease in mutation rate (Table 1), whereas only a twofold decrease of mutation rate in the polα(swi7-H4) polζΔ dinBΔ triple mutant than the polα(swi7-H4) single mutant (Table 2). It is also possible that Rad17 may promote another error-prone pathway, which facilitates the generation of deletion mutations. As shown in Figure 3, polα(swi7-H4) rad17.K118E accumulates fourfold less point mutations than the polα(swi7-H4) single mutant, whereas deletions of both dinB+ and polζ+ in polα(swi7-H4) reduce the point mutations 10-fold less than the polα(swi7-H4) single mutant. In this study, we only demonstrate the requirement of functional Rad17 for DinB to associate with chromatin for mutagenic synthesis. It is not yet known whether functional Rad17 is also required for Polζ to associate to chromatin for mutagenic synthesis. Polζ may be able to perform mutagenic synthesis to generate point mutations independent of the ability of Rad17 to associate with chromatin. Alternatively, rad17.K118E could be hypomorphic mutant in recruiting translesional polymerases.
The striking suppression of both deletion and point mutations in polα(swi7-H4) by rad17.K118E mutation suggest that Rad17, in addition to being involved in checkpoint to signal cell-cycle arrest, has other important physiological roles in activating translesion synthesis processes.
Checkpoint clamp-loader and checkpoint-clamp regulate the translesional polymerase DinB for mutagenic translesion synthesis.
Recruitment of DinB to chromatin requires the chromatin binding ability of Rad17 after genomic perturbation. In addition to the checkpoint-clamp-loader, Rad17, and the checkpoint-clamp, Rad3/Rad26 complex is thought to sense genomic perturbations of late S-phase and DNA damage independently from that of checkpoint-clamp/clamp-loader (Edwards et al. 1999; Michelson and Weinert 1999). It is not yet known whether Rad3/Rad26 is required in recruiting the translesional polymerases onto chromatin for mutagenic synthesis when replication is perturbed. Analysis of the effect of Rad3/Rad26 on the point mutation formation is hindered by the fact that DinB expression is not up-regulated in rad3Δ cells (Fig. 4).
The physical interactions between DinB with Hus1 and Rad1 suggest that the checkpoint-clamp might recruit and secure the DinB protein on chromatin for effective mutagenic translesion synthesis. Alternatively, the chromatin-bound checkpoint-clamp could somehow help DinB to replace the replicative polymerase or to stimulate the DNA synthetic activity of DinB. In vitro, proliferating cell nuclear antigen (PCNA), together with replication factor C (RFC) and replication protein A (RPA), stimulates the DNA synthesis activity of human Polκ(DinB) (Haracska et al. 2002). Given the predicted structural similarity between PCNA and the checkpoint-clamp, the physical interaction between DinB and the Rad1–Rad9–Hus1 checkpoint-clamp may be similar to that of the DinB–PCNA interaction in vitro, stimulating the DinB polymerase activity, or securely positioning DinB in the repair complex for mutagenic translesion synthesis.
Among the three fission yeast translesion synthesis polymerases analyzed in this study, DinB contributes most to the elevated mutation rate causing point mutations in cells, whereas Polζ contributes to a lesser extent when replication is perturbed (Table 2; Fig. 3). In vitro studies of human Polκ have shown that Polκ is a promiscuous extender of mispaired primer termini, and that both Polκ and Polζ can extend mispaired primer ends efficiently on undamaged DNA. These in vitro findings have led to a proposed model: Polκ or Polζ can readily incorporate a nucleotide to a mispaired primer terminus in undamaged DNA. Once the nucleotide is inserted, Polκ and Polζ will dissociate from the primer terminus to allow Polδ to restart DNA synthesis (Washington et al. 2002). Given the low processive nature of polymerase α, the semidisabled polymerase α in polα(swi7-H4) could be expected to cause a stalled replication fork at the semipermissive temperature, frequently dissociating from the primer-template, and increasing the chances of forming mispaired primer-ends. Extension of the mispaired ends by DinB and/or Polζ would result in an elevated mutation rate by accumulating primarily point mutations in cells.
How might mutations in cds1+ and rad17+ affect the deletion mutations in polα(swi7-H4)?
Cds1 and its budding yeast homolog Rad53 are thought to be essential in preventing accumulation of aberrant replication intermediates and in promoting replication restart (Desany et al. 1998; Lindsay et al. 1998; Lopes et al. 2001; Sogo et al. 2002). The cds1.T8A mutation in polα(swi7-H4) moderately compromised the checkpoint as shown by a faster advance of the cell cycle when compared to the polα(swi7-H4) single mutant (Fig. 1A). The polα(swi7-H4) cds1.T8A double mutant also had reduced Cds1 kinase activity. Furthermore, Cds1 autophosphorylation in this double mutant was compromised (Fig. 1B). Importantly, the cds1.T8A mutation in polα(swi7-H4) induced a hyper-mutation rate (Table 1), causing exceedingly high levels of deletion mutations (Fig. 2). Deletions are thought to be caused by accumulation of single-stranded gaps in the template, which are prone to double-strand breaks. Repair of double-strand breaks by recombination repair or by single-strand annealing would result in the deletion of a sequence flanked by homologous repeats (Paques and Haber 1999). In addition, a stalled replication fork induced by a semidisabled polymerase, such as in polα(swi7-H4), could also regress to form Holliday junction. A recent report has shown electron micrographic evidence that a budding yeast rad53 mutant accumulates single-stranded DNA and undergoes replication fork regression (Sogo et al. 2002). The cds1.T8A mutation may either positively or negatively affect the ability of repair factors in either error-free or error-prone processing the abnormal DNA structures and regressed replication forks. These could cause the accumulation of an exceedingly elevated level of deletion mutations in cells.
In contrast to the mutation in cds1+, mutations in rad17+ suppress the mutation rate and significantly reduce deletion mutations accumulated in polα(swi7-H4) (Table 1; Fig. 2). The rad17.K118E mutation in polα(swi7-H4) reduced the deletion mutations in cells from 194-fold higher than in wild-type cells to 11-fold, and point mutations from 16-fold higher than in wild-type cells to approximately fourfold (Fig. 2). The double mutant polα(swi7-H4) rad17.K118E has an intact checkpoint at the semipermissive temperature (Fig. 1A), and a slightly reduced level of Cds1 kinase activity (Fig. 1B). In the polα(swi7-H4) rad17.K118E double mutant, Cds1 is not autophosphorylated at the semipermissive temperature (Fig. 1B). These indicate that, although the checkpoint is functional in delaying the cell cycle transition and Cds1 is moderately activated in the polα(swi7-H4) rad17.K118E double mutant, some other biological responses are compromised. The reduction of deletion mutations in polα(swi7-H4) rad17.K118E double mutant suggests that the rad17.K118E mutation compromises some repair processes required to prevent the occurrence of deletion mutations in addition to its inability to recruit DinB and/or Polζ for mutagenic synthesis.
Conclusion
In response to genomic perturbations, the checkpoint is required to signal cell-cycle arrest to maintain genomic stability. Ironically, results of this study show that checkpoint activation can also induce genomic instability by activating the translesional polymerase for mutagenic translesion synthesis.
The mutagenic nature of translesional DNA polymerases operating on undamaged DNA is a troublesome source of genomic mutation. Knowledge of how these translesional polymerases are regulated is important for our understanding and, ultimately, prevention of genomic instability. Here, we have demonstrated that expression of the translesional polymerase DinB is regulated by checkpoint activation. Our results suggest that for DinB to associate with chromatin to perform mutagenic translesion synthesis in order to allow a stalled replication fork to restart, this process requires the ability of Rad17 to associate with chromatin. For DinB to be securely positioned on the chromatin, to enhance DinB polymerase activity, or to replace the replicative polymerase at the replication fork, an interaction between DinB and the checkpoint-clamp is required. Together, these findings suggest that mutagenic translesion synthesis by DinB and perhaps also some other translesional polymerases is a part of the checkpoint response. A model of how checkpoints respond to replication perturbations to arrest the cell cycle in order to maintain genomic stability and to activate translesional polymerases for mutagenic translesion synthesis is proposed (Fig. 6).
Figure 6.
A proposed model of how checkpoint responds to replication perturbations. A semidisabled DNA polymerase stalls the replication fork. Checkpoint sensors/transducers detect the perturbation and activate Cds1 and/or Chk1 to delay or arrest the cell cycle. Checkpoint activation also induces the expression of translesional polymerase DinB, perhaps also Polζ, which is recruited onto chromatin by interacting with checkpoint clamp to perform mutagenic translesion synthesis in order to restart the replication fork.
Materials and methods
General methods
All yeast media, YE5S, Edinburgh minimal medium, and 5-fluoroorotic acid (5-FOA) medium were prepared as described in Moreno et al. (1991) and Liu et al. (1999). Except where otherwise stated, all yeast strains were propagated at 25°C. All fission yeast genetic operations were performed as described in Gutz et al. (1974). For the disruption of polζ+ and the rad30+ domain of eso1+, a one-step gene disruption method was used, followed by PCR-mediated generation of the kan+ gene flanked by 80-bp segments from the 5′ and 3′ regions of the gene to be disrupted (Bahler et al. 1998). The DinB+ deletion strain was kindly provided by Dr. A.M. Carr (Genome Damage and Stability Centre, University of Sussex, UK). Epitope tagging of genes with either the myc- or HA-tag at their chromosomal loci was performed as described (Bahler et al. 1998). For Northern analysis, RNA was prepared from a 10-mL culture (5 × 107 cells) by using the RNeasy kit (Qiagen). [α-32P]-dATP-labeled probes were prepared by the random priming method. A PCR fragment of dinB+ was used as the probe (C-terminal 538-base DinB fragment). A DraI-EcoRV his3+ fragment was used as the loading control. Northern blot analysis was performed as described in Maniatis et al. (1982).
Mutation rate analysis
Mutation rates on ura4+ were calculated by fluctuation analysis by using the method of the median as previously described (Lea and Coulson 1949). The reported mutation rates are the average of values obtained from at least three independent experiments using sets of 11 independent cultures. Significance levels between strains were determined by using the Mann-Whitney rank test with Analyse-it, a plug-in statistical package for Microsoft Excel. Unless specified otherwise, all comparisons had a “significant” p level of p < 0.005.
Mutation spectra analysis
Genomic DNA was isolated from 5-FOAr colonies. The mutant ura4 gene was amplified, analyzed by 1.5% agarose gel electrophoresis, and classified as having either deletions or point mutations. The ura4 PCR products were also gel purified and sequenced to determine the mutations generated. Relevant genomic fragments were amplified by PCR. At least 100 5-FOAr colonies from each strain were analyzed for the size of ura4+-PCR product by agarose gel electrophoresis, and 20 ura4+-PCR products were sequenced. The PCR reactions included the ura4+ primers (P0, AAGCTTAGCTACAAATCCCAC; and P8, AAC GCCTAGGAAAACAAACGC). In addition to primer P0 and P8, the PCR products were sequenced using the following primers: P1, TTTCTTACCGTATTGTCCTAC; and P2, GGAC CCTATCTCTGTGTTATC.
Immunoprecipitation and immunoblotting
Cell lysates were prepared as in Kai et al. (2001) with the addition of 300 mM NaCl and 1% TX-100 to the lysis buffer. Before immunoprecipitation, 100 U of DNaseI was incubated with each reaction, and immunoprecipitation was performed as described in Kai et al. (2001).
Cds1 kinase assay and chromatin fractionation assay
Cds1 kinase assays and chromatin fractionation of myc-tagged DinB was performed as in Kai et al. (2001). Cds1 auto-phosphorylation was analyzed as in Tanaka et al. (2001).
Flow cytometry analysis
Cells were harvested, washed in water, and fixed in 70% ethanol. DNA content was measured by using a FACScan system, CellFIT cell cycle analysis, and LYSISII software (Becton Dickinson).
Acknowledgments
We thank members of the Wang lab for helpful discussion and Karlene A. Cimprich for critical reading of the manuscript, Antony M. Carr for dinB deletion and rad17 mutant strains, and Paul Russell for the cds1.T8A mutant strain. This study was supported by grants from National Cancer Institute of National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL twang@pmgm2.stanford.edu; FAX: (650) 725-6902.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1043203.
References
- Al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bhaumik D, Wang TSF. Mutational effect of fission yeast Polα on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Caspari T, Carr AM. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochemie. 1999;81:173–181. doi: 10.1016/s0300-9084(99)80050-9. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hoffmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;74:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Scheits JL, Amin NS, Lau PJ, Myung K, Kolodner RD. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol Cell. 2000;6:593–603. doi: 10.1016/s1097-2765(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PJ, Bentley NJ, Carr AM. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: Preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington D.C.: ASM Press; 1995. [Google Scholar]
- Friedberg EC, Feaver WJ, Gerlach VL. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci. 2000;97:5681–5683. doi: 10.1073/pnas.120152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- Griffiths DJF, Barbet NC, McCready S, Lehmann AR, Carr AM. Fission yeast rad17: A homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths D, Uchiyama M, Nurse P, Wang TSF. A novel allele of the chromatin-bound fission yeast checkpoint protein Rad17 separates the DNA structure checkpoints. J Cell Sci. 2000;113:1075–1088. doi: 10.1242/jcs.113.6.1075. [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of genetics 1. New York, NY: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- Kai M, Tanaka H, Wang TSF. Fission yeast Rad17 binds to chromatin in response to replication arrest or DNA damage. Mol Cell Biol. 2001;21:3289–3301. doi: 10.1128/MCB.21.10.3289-3301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJF, Edwards R, Murray JM, Christensen PU, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in S. pombe. Genes & Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu VF, Bhaumik D, Wang TSF. Mutator phenotype induced by aberrant replication. Mol Cell Biol. 1999;19:1126–1135. doi: 10.1128/mcb.19.2.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: A laboratory manual. Cold Spring Harbor, New York, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Masutani C, Araki M, Yamada A, Kusumoto T, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson R, Weinert T. Sensor-less checkpoint activation? Na Cell Biol. 1999;1:177–178. doi: 10.1038/15614. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle CD. Thymine–thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- O’Connell MJ, Walworth NC, Carr AM. The G2-phase DNA damage checkpoint. Trends Cell Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J Biol Chem. 2000a;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes & Dev. 2000b;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Opperman T, Murli S, Smith BT, Walker GC. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Armour CD, Hartwell LH. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics. 1998;150:75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Chk1 and Cds1: Linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Sutton MD, Smith BT, Godoy VG, Walker GC. The SOS response: Recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu Rev Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa Y, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Boddy MN, Chen XB, McGowan CH, Russell P. Threonine-11, phosphorylated by Rad3 and ATM in vitro, is required for activation of fission yeast checkpoint kinase Cds1. Mol Cell Biol. 2001;21:3398–3404. doi: 10.1128/MCB.21.10.3398-3404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Wang Z. Translesion synthesis by the UmuC family of DNA polymerases. Mutat Res. 2001;486:59–70. doi: 10.1016/s0921-8777(01)00089-1. [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase eta. Proc Natl Acad Sci. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes & Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes & Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]