Abstract
Haploinsufficiency of Tbx1 is likely a major determinant of cardiac and craniofacial birth defects associated with DiGeorge syndrome. Although mice deficient in Tbx1 exhibit pharyngeal and aortic arch defects, the developmental program and mechanisms through which Tbx1 functions are relatively unknown. We identified a single cis-element upstream of Tbx1 that recognized winged helix/forkhead box (Fox)-containing transcription factors and was essential for regulation of Tbx1 transcription in the pharyngeal endoderm and head mesenchyme. The Tbx1 regulatory region was responsive to signaling by Sonic hedgehog (Shh) in vivo. We show that Shh is necessary for aortic arch development, similar to Tbx1, and is also required for expression of Foxa2 and Foxc2 in the pharyngeal endoderm and head mesenchyme, respectively. Foxa2, Foxc1, or Foxc2 could bind and activate transcription through the critical cis-element upstream of Tbx1, and Foxc proteins were required, within their expression domains, for Tbx1 transcription in vivo. We propose that Tbx1 is a direct transcriptional target of Fox proteins and that Fox proteins may serve an intermediary role in Shh regulation of Tbx1.
Keywords: Tbx1, Shh, Fox, DiGeorge syndrome
The 22q11.2 deletion syndrome (22q11DS) is the most frequent chromosomal microdeletion syndrome in humans and is typically characterized by a 3-Mb heterozygous deletion, congenital cardiac outflow tract, aortic arch, and craniofacial defects (Scambler 2000; Lindsay 2001; Yamagishi 2002). The embryologic basis for this disease, also known as DiGeorge syndrome, resides in tissues derived from, or cells traversing through, the pharyngeal arches. The pharyngeal arches are bilaterally symmetric structures that arise in a segmental fashion along the anterior–posterior (AP) axis. They are precisely patterned by clustered homeobox (Hox) genes that establish unique identity to each arch based on its AP positioning (for review, see Schilling 1997). Numerous epithelial–mesenchymal interactions are necessary for appropriate segmentation and outgrowth of the pharyngeal arch in a manner similar to the developing limb bud, where Sonic hedgehog (Shh), bone morphogenetic proteins (BMPs), fibroblast growth factors (FGFs), and Wnts regulate patterning along all three embryonic axes (for review, see Martin 2001).
Each pharyngeal arch consists of an aortic arch artery and a mesodermally derived core, covered externally and internally by ectoderm and endoderm-derived epithelium, respectively (Sadler 2000). The bilaterally symmetric aortic arch arteries undergo extensive remodeling to ultimately form the mature aortic arch and pulmonary artery, whereas the mesodermal core gives rise to facial and neck muscles and the cardiac outflow tract. The endoderm-derived epithelial layer is essential for segmentation of the pharyngeal arches and contributes to craniofacial organs, including the thymus and parathyroid glands. Neural crest cells migrate from the caudal hindbrain to each pharyngeal arch, surrounding the aortic arch arteries and central core of mesodermal cells, with some cells proceeding to populate the cardiac outflow tract (conotruncus). The neural-crest-derived mesenchyme contributes to skeletal structures of the face and is important in patterning the aortic arch and septation of the conotruncus (Kirby and Waldo 1995). Reciprocal interactions between the various cell types are essential for proper development of the pharyngeal arch derivatives.
Recent evidence in mice indicates that haploinsufficiency of Tbx1, a T-box transcription factor that is expressed in the developing pharyngeal arches (Chapman et al. 1996; Garg et al. 2001), may be a major contributor to aortic arch defects in the 22q11 deletion phenotype. Mice harboring a hemizygous deletion of part of the mouse equivalent of 22q11.2 have some features of 22q11DS, including aortic arch defects and occasional conotruncal and thymic defects (Lindsay et al. 1999; Merscher et al. 2001). A subset of mice heterozygous for Tbx1 display defects in aortic arch patterning that are reminiscent of defects seen in 22q11DS (Jerome and Papaionnou 2001; Lindsay et al. 2001; Merscher et al. 2001). Furthermore, aortic arch defects observed in mouse chromosomal deletions syntenic to 22q11 can be rescued by a transgene encompassing Tbx1 (Lindsay et al. 2001; Merscher et al. 2001). Mice homozygous-null for Tbx1 display additional features of 22q11DS, including pharyngeal arch segmentation defects, strengthening a role for Tbx1 in this syndrome (Jerome and Papaionnou 2001; Vitelli et al. 2002a).
Tbx1 belongs to a large family of transcription factors that shares an evolutionarily conserved T-box first described in the brachyury (T) protein that is necessary for mesoderm formation (Bollag et al. 1994). The T-box is necessary for DNA-binding and can mediate combinatorial interactions with other transcriptional regulators (for review, see Papaionnou and Silver 1998; Smith 1999). Tbx1 is expressed in discrete domains, including the pharyngeal endoderm and the pharyngeal mesoderm, and broadly in the head mesenchyme (Chapman et al. 1996; Garg et al. 2001). It is notable that Tbx1 is excluded from the neural-crest-derived mesenchyme of the pharyngeal arches, although signaling from the epithelial and mesodermal layers can affect differentiation of the adjacent neural crest cells. Recent evidence suggests that Tbx1 may regulate fibroblast growth factor 8 (Fgf8) in the endoderm, which may signal to adjacent neural crest cells and regulate aortic arch development (Abu-Issa et al. 2002; Frank et al. 2002; Vitelli et al. 2002b).
We previously suggested that Tbx1 responds to signals initiated by the secreted morphogen Shh. Shh mutant mice die soon after birth with severe craniofacial defects (Chiang et al. 1996; Helms et al. 1997) and fail to maintain Tbx1 expression (Garg et al. 2001). Consistent with a role for Shh in regulation of Tbx1, chick embryos exposed to Shh-soaked beads responded by up-regulating Tbx1 expression in the pharyngeal arches (Garg et al. 2001). However, the mechanism through which this phenomenon occurred was unknown. Here, we describe a genomic regulatory region that controls the in vivo expression of Tbx1 and is responsive to Shh signaling. We demonstrate that a single cis-element that recognizes winged helix/forkhead box (Fox)-containing transcription factors (Kaufmann and Knochel 1996; Kaestner et al. 2000) is essential for regulation of Tbx1 transcription in the pharyngeal endoderm and head mesenchyme. Foxa2, required for endoderm development (Ang and Rossant 1994; Weinstein et al. 1994), and Foxc1 or Foxc2, required for head mesenchyme and aortic arch formation (Iida et al. 1997; Winnier et al. 1997, 1999; Kume et al. 1998, 2001), bind and activate Tbx1 transcription in their respective subdomains through this regulatory element. We show that Shh is necessary for aortic arch development, similar to Tbx1, and is also required for expression of Foxa2 and Foxc2 in the pharyngeal endoderm and head mesenchyme, respectively. These studies reveal a cascade of events that culminates in transcription of Tbx1 and establish, to our knowledge, the first known direct target of Foxc proteins during development.
Results
Separable enhancers direct tissue-specific expression of Tbx1
To define the developmental program in which Tbx1 functions, we searched for the cis-regulatory elements that control Tbx1 expression during development. We isolated a mouse bacterial artificial clone (BAC) containing genomic sequence 5′ of Tbx1 and subcloned a fragment of DNA extending from the 5′-untranslated region (UTR) to −14.3 kb (12.8 kb in length) upstream of a lacZ reporter gene (Fig. 1a, construct 1). Mouse embryos harboring this transgene showed X-gal staining in the mesodermal core and endodermal epithelium of pharyngeal arches, as well as in the head mesenchyme surrounding the dorsal aortas (Fig. 1d,e). This pattern accurately recapitulated the expression of endogenous Tbx1 transcripts (Fig. 1b,c).
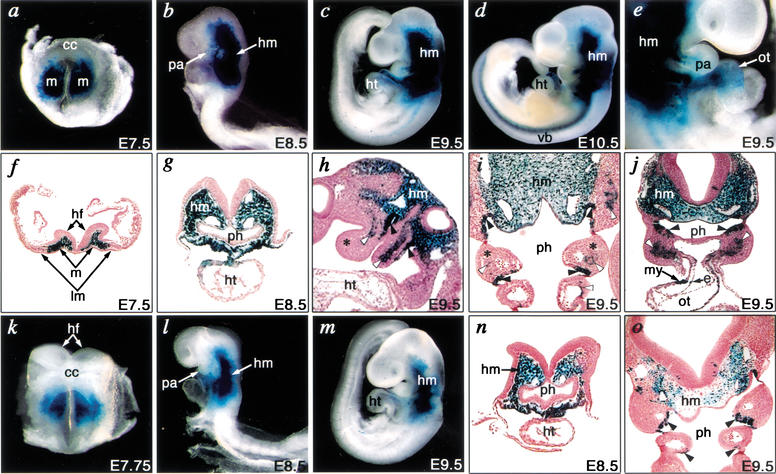
Figure 1.
Tbx1 expression is controlled by separable pharyngeal endoderm and mesoderm regulatory regions in transgenic mice. (a) Genomic organization of the 5′ mouse Tbx1 locus and flanking region. Boxes indicate exons, and translation start site (ATG) is designated as nucleotide number zero. Construct number is indicated on the left, and the corresponding expression pattern of lacZ at E9.5 is summarized on the right. The far-right column indicates the fraction of F0 transgenic embryos with Tbx1-like lacZ expression/lacZ gene positive embryos. (b,c) Endogenous expression of Tbx1 transcripts is shown by whole-mount RNA in situ hybridization in E9.5 mouse embryos. (d–k) Right lateral views of representative embryos obtained with each construct (indicated in upper-right corner of each panel). (c,e,g,i,k) Higher magnification of the pharyngeal arch of embryos in upper panels. Expression of lacZ in embryos with construct 1 (d,e) recapitulated endogenous Tbx1 expression (b,c). LacZ expression in the head mesenchyme (hm) and pharyngeal endoderm (white arrowheads) was detectable in each embryo with construct 1 (e), 3 (g), 4 (i), and 6 (k), whereas that in the pharyngeal mesoderm (black arrowheads) and cardiac outflow tract (ot) was only in embryos with construct 1 (e) or 3 (g). lacZ expression in limb buds (lb) in panel h represents ectopic expression. h, head; ht, heart.
To localize the regulatory elements responsible for embryonic expression of Tbx1, we constructed a series of deletions of the 12.8-kb genomic fragment and analyzed their ability to direct lacZ expression from a heterologous promoter (hsp68) in F0 transgenic mice at embryonic day 9.5 (E9.5). Comparison of sequence upstream of mouse and human TBX1 within a 6.4-kb fragment (construct 3) that was sufficient for appropriate expression (Fig. 1f,g) revealed sequence conservation in four regions spanning 1.5 kb (−12.8 to −14.3 kb). This 1.5-kb fragment (construct 4) directed lacZ expression in pharyngeal arch endoderm and head mesenchyme, but not in pharyngeal mesoderm (Fig. 1h,i). Further deletions revealed that the critical cis-element(s) necessary for Tbx1 expression in the pharyngeal endoderm and head mesenchyme lay between −14.3 and −13.2 kb (Fig. 1j,k).
Enhancer-dependent temporospatial expression of Tbx1
The temporospatial expression pattern of Tbx1 during early embryogenesis was determined using stable transgenic mouse lines bearing lacZ under control of the 12.8-kb (construct 1) or 1.1-kb (construct 6) upstream fragments described above. At E7.5, the 12.8-kb transgene directed lacZ expression in the paraxial mesodermal cells that give rise to cranial mesenchyme, but not in the lateral plate mesoderm or cardiac crescent that forms the heart tube (Fig. 2a,f). Slightly later (E8.5), lacZ expression was observed in a segment of the cranial mesoderm including the first pharyngeal arch mesoderm (Fig. 2b,g). At E9.5, the transgene directed high levels of lacZ expression in the mesoderm-derived head mesenchyme and the mesodermal core of the pharyngeal arches (Fig. 2c,h–j). LacZ expression in pharyngeal endoderm became detectable at this stage and was pronounced in the pharyngeal pouches. Expression was not detectable in pharyngeal arch mesenchyme derived from neural crest cells, consistent with the endogenous Tbx1 expression previously shown by RNA in situ hybridization. At E10.5, lacZ expression was maintained in the head mesenchyme, pharyngeal mesoderm, and endoderm, and began to extend to the precursors of the vertebral column (Fig. 2d). Although expression of Tbx1 in the developing heart had not been detectable by mRNA expression analyses, a recent study using mice harboring the lacZ gene targeted into the Tbx1 locus demonstrated Tbx1 expression in the cardiac outflow tract between E9.5 and E12.5 (Vitelli et al. 2002a,b). LacZ expression under control of the 12.8-kb fragment was detectable in the myocardial and endocardial layer of the outflow tract between E8.5 and E10.5 (Fig. 2e,j), indicating that Tbx1 may be expressed in the recently identified cells of the anterior heart tube that are derived from pharyngeal mesoderm (Kelly et al. 2001; Mjaatvedt et al. 2001; Waldo et al. 2001).
Figure 2.
Developmental regulation of Tbx1 expression by tissue-specific enhancers. Embryos from a stable transgenic line harboring the 12.8-kb fragment (construct 1) were analyzed at various times during mouse embryogenesis. (a–d) Whole-mount photographs of embryos from E7.5 to E10.5. (e) Right lateral view of embryo focusing on the pharyngeal arch and heart at E9.5. (f–j) Transverse (f,g,j), sagittal (h), and frontal (i) sections counterstained by Nuclear Fast Red from E7.5 to E9.5. (a,f) lacZ was expressed in mesoderm cells (m) that give rise to cranial mesenchyme, but not in the cardiac crescent (cc) or lateral plate mesoderm (lm) at E7.5. (b,g) Expression of lacZ was detectable in the head mesenchyme (hm) and pharyngeal arch (pa) at E8.5. (c,h,i,j) lacZ expression was detectable in head mesenchyme, pharyngeal arch mesoderm (white arrowheads), and endoderm (black arrowheads), but not in neural-crest-derived mesenchyme (asterisks) at E9.5. (d) Expression of lacZ extended to the primordia of vertebral bodies (vb). (e,j) lacZ was expressed in both myocardial (my) and endocardial (e) layers of the cardiac outflow tract (ot) at E9.5. (k–m) Whole-mount photographs of embryos from a stable transgenic line harboring the 1.1-kb fragment (construct 6) at E7.75 to E9.5. (n,o) Transverse (n) or frontal (o) sections of l or m, respectively, counterstained by Nuclear Fast Red. (k) lacZ expression in mesoderm cells was observed similar to panel a at E7.75. (l,n) lacZ was detectable in head mesoderm as in b and g, but excluded from the pharyngeal arch at E8.5. (m,o) Expression of lacZ in head mesenchyme and pharyngeal endoderm was detectable similar to c, i, and j, but expression in pharyngeal mesoderm was absent. hf, head fold; ht, heart; ph, pharynx.
A stable transgenic mouse line harboring the 1.1-kb enhancer (construct 6) showed lacZ expression in the head mesenchyme from E8.0 through E10.5, although it was relatively weaker than that of transgenic mice with the 12.8-kb fragment (Fig. 2k–o). Differences in expression level could be secondary to positional effects of transgene insertion or other elements that further enhance expression. The 1.1-kb enhancer also directed lacZ expression in the pharyngeal arch endoderm from E9.5, similar to the 12.8-kb fragment, but excluded pharyngeal arch mesoderm and cardiac outflow tract expression (Fig. 2l,m,o), indicating that separable enhancers direct distinct domains of Tbx1 expression.
Tbx1 enhancer is responsive to Shh signaling
We have previously reported regulation of Tbx1 by Shh in mouse and chick embryos (Garg et al. 2001). Shh-null mice die soon after birth and have numerous craniofacial defects (Chiang et al. 1996; Helms et al. 1997) that are distinct but overlapping with defects in Tbx1 mutants. To determine whether the identified enhancer of Tbx1 was dependent on Shh signaling, we examined activity of the Tbx1–lacZ transgene in the Shh-null background. At E9.25, lacZ expression was relatively normal in the pharyngeal arches and head mesenchyme of Shh-null mutant embryos compared with wild-type embryos (Fig. 3a,b). By E9.5, lacZ expression was detectable but down-regulated in pharyngeal arches and the head mesenchyme of Shh mutant embryos (Fig. 3c–f). Only 1 d later (E10.5), lacZ expression was nearly undetectable in Shh mutant embryos, consistent with our previous results from Tbx1 mRNA in situ hybridization (Garg et al. 2001). These results suggest that the Tbx1 enhancer responds to Shh signaling and promotes maintenance, rather than induction, of Tbx1 expression in a temporo–spatial fashion.
Figure 3.
The Tbx1 enhancer is dependent on Shh signaling in a temporo–spatial fashion. (a,b) X-gal staining of wild-type (Shh+/+) and Shh mutant (Shh−/−/) embryos harboring the 12.8-kb Tbx1–lacZ transgene at E9.25 showed relatively normal lacZ expression in the pharyngeal arches and head mesenchyme (hm) of Shh mutant embryos compared with wild-type embryos. (c,d) LacZ expression in wild-type and Shh mutant embryos at E9.5. (e,f) Higher magnification photographs of c and d focusing on pharyngeal arches show down-regulation of lacZ expression in the head mesenchyme and first (I), third (III), and fourth (IV) pharyngeal arches of Shh mutant embryos compared with wild-type embryos. lacZ expression in the second pharyngeal arch (II) was also down-regulated, but expression was still detected in the mesoderm of the second arch of Shh mutant embryos. ht, heart.
A forkhead-transcription-factor-binding element is necessary for Tbx1 transcription in pharyngeal endoderm and head mesenchyme
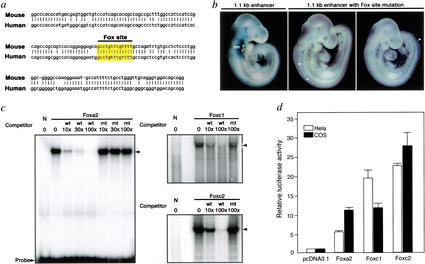
We inspected the 1.1-kb enhancer for binding sites of transcription factors using the MatInspector V.2.2 based on TRANSFAC 4.0 (Quandt et al. 1995). We did not identify a consensus binding site for Gli proteins that might mediate transcriptional activities directly downstream of Shh signaling (Sasaki et al. 1997), suggesting that the regulation of the Tbx1 enhancer by Shh may be through an intermediate step. Alignment of the 1.1-kb mouse Tbx1 enhancer with the corresponding human TBX1 upstream sequence revealed conservation of several potential transcription-factor-binding sites. In particular, a consensus binding site for the winged helix/forkhead transcription factor family (GCCTGTTTGTTTT) was completely conserved across species (Fig. 4a). Members of the winged helix/forkhead class of transcription factors, also known as Fox proteins (Kaestner et al. 2000), serve as critical regulators during embryogenesis, tumorigenesis, and in maintenance of differentiation (Kaufmann and Knochel 1996). To determine if the Fox-binding site was required for Tbx1 expression in vivo, we mutated this site in the context of the 1.1-kb enhancer and generated transgenic embryos. LacZ expression in pharyngeal endoderm and head mesenchyme was abolished in 6/6 embryos harboring the mutant transgene, although ectopic expression of lacZ was observed in some embryos (Fig. 4b). This result identified Tbx1 as a direct transcriptional target of Fox transcription factors during development in vivo.
Figure 4.
A consensus Fox-binding site is essential for regulation of Tbx1. (a) One-hundred percent cross-species conservation of the consensus Fox-binding site at −13,435/−13,423 bp in the 1.1-kb enhancer of Tbx1 is indicated. (b) Mutation of Fox site abolished head mesenchyme (hm) and pharyngeal endoderm (asterisks) enhancer activity in 6/6 lacZ-positive embryos. Ectopic lacZ staining (arrowheads) was observed in some embryos. (c) Electrophoretic gel mobility shift assays (EMSA) revealed DNA–protein complexes of in vitro translated Foxa2, Foxc1, or Foxc2 with the Fox cis-element in the Tbx1 enhancer. The interaction was competed specifically by excess unlabeled wild-type (wt) oligonucleotide, but not by a mutant (mt) oligonucleotide. Fold-excess of competitor DNA compared with labeled probe is shown. No DNA–protein complex (in lane N) was observed with lysate alone. (d) Luciferase expression under control of the Tbx1 genomic DNA fragment shown in panel a after cotransfection of luciferase reporter and Foxa2, Foxc1, or Foxc2 expression construct or empty vector (pcDNA3.1). Results are presented as relative luciferase activity, corrected for transfection efficiency. The control (pcDNA3.1) transfection value is set at 1.
Tissue-specific forkhead proteins bind and activate Tbx1 transcription through a common cis-acting element
Of the numerous Fox factors, Foxc1 and Foxc2 (formerly Mf1 and Mfh1, respectively) are closely related Fox proteins with virtually identical DNA-binding domains that are expressed in the mesoderm-derived head mesenchyme and play redundant roles in cardiovascular development (Iida et al. 1997; Winnier et al. 1997, 1999; Kume et al. 1998, 2001). Embryos lacking either Foxc1 or Foxc2, and most compound heterozygotes, die perinatally from aortic arch defects, including interruption of the aortic arch (IAA; Winnier et al. 1999; Kume et al. 2001), reminiscent of mice heterozygous for a Tbx1 mutation (Jerome and Papaionnou 2001; Lindsay et al. 2001; Merscher et al. 2001). In contrast, Foxa proteins, also known as hepatocyte nuclear factors (HNF)-3, are expressed in the pharyngeal endoderm (Kaufmann and Knochel 1996), and one member, Foxa2 (HNF-3β), is essential for endoderm development (Ang and Rossant 1994; Weinstein et al. 1994). Based on these data, we hypothesized that Foxa2 might regulate expression of Tbx1 in the pharyngeal endoderm, whereas Foxc1 and Foxc2 may regulate expression of Tbx1 in the head mesenchyme through a common cis-regulatory element.
To determine if the conserved Fox-binding site was capable of binding Foxa2, we performed electrophoretic mobility shift assays (EMSA) with 32P-labeled oligonucleotides containing the cis-element. Foxa2 protein specifically shifted this element, and DNA binding was effectively competed by an excess of unlabeled cognate competitor but not by a mutated oligonucleotide, suggesting that Foxa2 specifically interacts with the Fox consensus site in the Tbx1 enhancer (Fig. 4c). Similar studies using Foxc1 or Foxc2 protein also demonstrated interaction with this site with varying affinities (Fig. 4c).
To test whether Fox proteins could not only bind, but also activate transcription through this site, we transiently transfected HeLa cells with a luciferase reporter cloned downstream of the Fox-binding element (FBE) in the 1.1-kb Tbx1 enhancer. Transfection with expression vectors of Foxa2, Foxc1, or Foxc2 activated this reporter by ∼5-, ∼20-, or ∼25-fold, respectively (Fig. 4d). Similar results were obtained in COS-7 cells (Fig. 4d), suggesting that Foxa2, Foxc1, and Foxc2 can bind and activate transcription through the consensus Fox cis-element upstream of Tbx1 in vitro.
Foxc1 and Foxc2 are required for normal Tbx1 expression in vivo
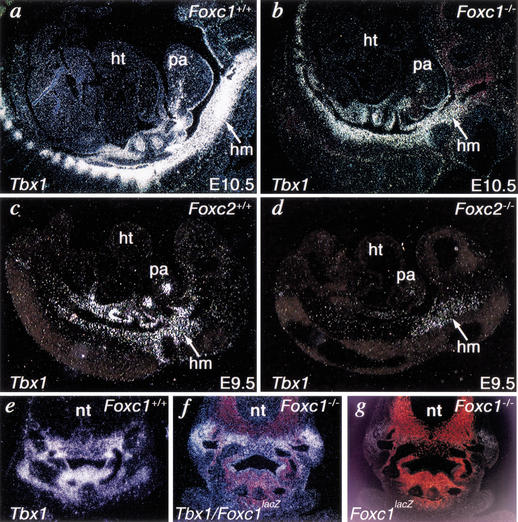
Unlike Foxa2 mutants, Foxc1-null or Foxc2-null embryos survive until E18.5 (Iida et al. 1997; Winnier et al. 1997; Kume et al. 1998), allowing us to test whether these factors are involved in regulation of Tbx1 in vivo. The expression of Tbx1 was partially down-regulated in either Foxc1-null or Foxc2-null embryos at E9.5–E10.5 (Fig. 5a–d), suggesting that Foxc1 and Foxc2 may regulate Tbx1 in a dose-dependent and redundant fashion. Comparison of the domains of Foxc1 expression, represented by lacZ-positive cells (red), and Tbx1 expression (white) in wild-type or Foxc1 mutants revealed that Tbx1 expression in Foxc1 mutants was only affected in areas in which Foxc1 and Tbx1 were coexpressed, consistent with Foxc regulation of Tbx1 (Fig. 5e–g). It is worth noting that cell numbers in Foxc mutants are comparable to wild type as no defect in cell death or proliferation has been found (Winnier et al. 1999; T. Kume, unpubl.). Mice homozygous-null for both Foxc1 and Foxc2 die at E9.5–E10.5 from abnormal development of the vasculature and pharyngeal arches (Kume et al. 2001), precluding formal examination of Tbx1 expression in Foxc1/Foxc2 double homozygous mutants.
Figure 5.
Foxc proteins regulate Tbx1 expression in vivo. (a–d) Radioactive in situ hybridization on sagittal sections of E9.5–E10.5 mouse embryos showed that Tbx1 was expressed, but partially down-regulated in the head mesenchyme (hm) of Foxc1 or Foxc2 mutant (Foxc1−/− or Foxc2−/−) embryos compared with wild-type (Foxc1+/+ or Foxc2+/+) embryos. (e–g) Transverse section of wild-type or Foxc1 mutants at the pharyngeal arch level. Tbx1 expression (white) in Foxc1-null embryo was down-regulated (f) compared with wild type (e) in the subdomain corresponding to Foxc1 expression, as marked by lacZ expression under control of the endogenous Foxc1 promoter (red in g). Tbx1 expression and lacZ expression are overlaid in f. ht, heart; pa, pharyngeal arch; nt, neural tube.
Foxa2 is dependent on Shh signaling in the pharyngeal endoderm
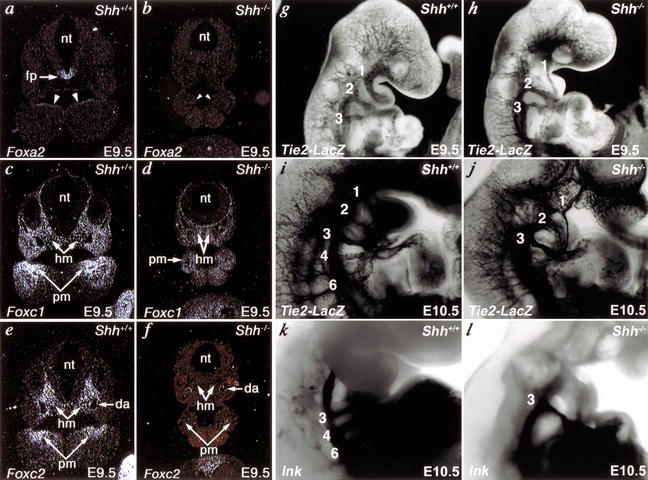
Previous studies demonstrated that the expression of Foxa2 was down-regulated in the neural tube floor plate of Shh-null mice from E8.5, suggesting that Shh signaling may be required for maintenance, but not induction, of Foxa2 expression (Chiang et al. 1996; Kaufmann and Knochel 1996; Sasaki et al. 1997). Identification of a Gli-dependent floor plate enhancer upstream of Foxa2 further established Foxa2 as a direct transcriptional target of Shh signaling (Sasaki et al. 1997). We specifically examined expression of Foxa2 in Shh-null pharyngeal endoderm and found that Foxa2 was dependent on Shh signaling in this domain as well as in the neural tube floor plate (Fig. 6a,b). The early demise of Foxa2 mutants prevented formal examination of Tbx1 expression in pharyngeal arches of Foxa2 mutants. The down-regulation of Foxa2 in Shh mutant endoderm is consistent with Foxa2 involvement in the maintenance of endodermal Tbx1 expression by Shh.
Figure 6.
Shh regulates Fox proteins and aortic arch development. (a,b) Radioactive in situ hybridization on transverse sections of E9.5 mouse embryos shows that Foxa2 is down-regulated in floor plate (fp) of neural tube (nt) and pharyngeal endoderm (arrowheads) of Shh−/− embryos compared with wild-type (Shh+/+) embryos. (c,d) Foxc1 was expressed in the head mesenchyme (hm) and pharyngeal arch mesenchyme (pm) of Shh mutant embryos at relatively low levels compared with wild-type embryos. (e,f) Foxc2 was undetectable in the head mesenchyme and pharyngeal arch mesenchyme of Shh mutant embryos compared with wild-type embryos. Foxc2 expression in dorsal aortae (da) was normal in Shh mutant embryos. a, c, and e are serial sections from a wild-type embryo, and b, d, and f are serial sections from an Shh mutant embryo. (g–l), Aortic arch defects in Shh mutants. lacZ driven by the Tie2 promoter marked the vasculature and revealed a small first aortic arch artery at E9.5 (h) and absence of the fourth and sixth arch arteries at E10.5 (j) compared with wild type (g,i). India ink injection demonstrated similar absence of the fourth and sixth arch arteries. Arch arteries are indicated numerically. Right lateral views are shown with close-ups of the pharyngeal arch region in i–l.
Shh is required for Foxc2 expression and aortic arch development
Based on our observation that Foxc proteins directly regulate the Shh-responsive Tbx1 enhancer, we asked whether Foxc1 or Foxc2 might serve as a transcriptional link between Shh signaling and Tbx1 expression. To test this idea, we examined whether Foxc1 or Foxc2 expression was dependent on Shh signaling. Foxc2 expression was undetectable, whereas Foxc1 expression was detectable, although somewhat diminished, in the pharyngeal arches of Shh mutants at E9.5 (Fig. 6c–f). Because fewer cells are present in the Shh mutant pharyngeal arch, the apparent Foxc1 down-regulation may be secondary to alterations in cell number; however, the complete absence of Foxc2 expression suggests a requirement of Shh signaling for Foxc2 activation.
Because our data supported a model in which Fox proteins mediate a Shh signaling cascade resulting in maintenance of Tbx1 expression, we hypothesized that Shh mutants might have aortic arch defects similar to Tbx1 mutants. To better detail the developing aortic arch arteries, we crossed mice containing a lacZ reporter downstream of the endothelial-specific Tie2 promoter (Schlaeger et al. 1997) into the Shh-null background. At E9.5, the first, second, and third arch arteries were apparent in Shh mutant and wild-type embryos (Fig. 6g,h), although the first arch artery was smaller in the absence of Shh. The first and second arch arteries normally begin to regress by E10.5, at which time the fourth and sixth arch arteries are visible (Fig. 6i). In contrast, embryos lacking Shh had only a third aortic arch artery at this stage with no evidence of a fourth or sixth arch artery on the left or right side (Fig. 6j). This finding was confirmed by injection of India ink into the cardiovascular system to delineate the aortic arch, which also indicated the isolated presence of the third arch artery (Fig. 6k,l). Tbx1 heterozygotes have a diminutive or absent fourth arch artery at this stage, consistent with a common role for Shh and Tbx1 in aortic arch patterning and development.
Discussion
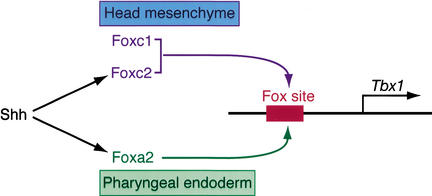
The pharyngeal arches are highly susceptible to perturbation as reflected by the numerous congenital syndromes affecting their derivatives. The close relationship of human cardiac malformations with pharyngeal arch derivatives reflects, in part, their common ontogeny. The signaling cascade involving Shh and Fox proteins that culminates on the regulatory apparatus of Tbx1, as revealed here (Fig. 7), provides an understanding of the events involved in development of the pharyngeal and aortic arch derivatives and some features of 22q11DS.
Figure 7.
A proposed model for molecular regulation of Tbx1. Our data support a model in which Tbx1 is directly regulated by Foxc1 and Foxc2 in the head mesenchyme, and Foxa2 in the pharyngeal endoderm, through a common Fox-binding site upstream of Tbx1. Shh signaling functions to maintain the expression of Foxa2 and Foxc2, which subsequently activate Tbx1 expression.
Transcriptional regulation of Tbx1 by Fox proteins
We found that a single Fox-binding element is essential for expression of Tbx1 in the head mesenchyme and in the pharyngeal endoderm. We propose that a common Fox-binding element can interpret transcriptional messages from Foxa or Foxc proteins to activate Tbx1 transcription in either the endoderm or head mesenchyme, respectively. Although Foxa and Foxc proteins have somewhat divergent forkhead DNA-binding domains and likely have varying affinities for the conserved Fox-binding site, it is interesting that a single cis-element translates information for both spatial domains of Tbx1 expression. It is possible that relative affinities for the binding element dictate onset of gene expression in the mesenchyme or endoderm, analogous to recent evidence in Caenorhabditis elegans, suggesting that variable affinity of the Foxa2 ortholog PHA-4 for distinct forkhead regulatory elements predicts activation of target genes (Gaudet and Mango 2002). Although the endoderm and mesenchyme enhancers are not separable, the region regulating mesoderm expression of Tbx1 appears to be under separate transcriptional regulation. Foxa proteins are inherently capable of initiating chromatin relaxation events (Cirillo et al. 2002) in collaboration with the zinc-finger transcription factor GATA4, suggesting a general mechanism for transcriptional activation by Fox proteins. It will be interesting to determine whether Foxa2 similarly cooperates with GATA3 to regulate Tbx1, as GATA3 is coexpressed in the pharyngeal endoderm (Lim et al. 2000) and is linked to a second DiGeorge syndrome locus on 10p (Gottlieb et al. 1998).
Foxc1 and Foxc2 are redundant transcription factors that are expressed in the head mesenchyme and mesenchyme surrounding the aortic arch arteries. The observation that mice lacking any two of the four alleles of Foxc1 or Foxc2 display aortic arch malformations (Winnier et al. 1999; Kume et al. 2001) strikingly similar to those of Tbx1 heterozygotes (Jerome and Papaionnou 2001; Lindsay et al. 2001; Merscher et al. 2001), including interruption of the aortic arch, is consistent with our data that Foxc proteins and Tbx1 function in a common program. To our knowledge, Tbx1 is the first direct target gene identified for Foxc1 or Foxc2 during development, and the down-regulation of Tbx1 in the mesenchyme of Foxc1−/− or Foxc2−/− embryos by at least 50% provides a potential mechanism for the aortic arch defects observed in Foxc mutants.
Fox proteins as mediators of Shh signaling
Our in vitro and in vivo studies demonstrate that one mechanism by which Shh signaling may be linked to Tbx1 transcription is through members of the Fox family of transcription factors. Shh signals through the transmembrane receptor Patched, which results in transcriptional activation of target genes by the Gli family of DNA-binding proteins (for review, see McMahon 2000). Shh is a direct upstream regulator of a Gli-binding site 5′ of Foxa2 that is necessary for Foxa2 expression in the floor plate of the notochord (Sasaki et al. 1997). Shh is also required for Foxa2 expression in the pharyngeal endoderm, as shown here, supporting an intermediary role for Foxa2 in Tbx1 regulation. The ability of Foxa2 to bind and activate transcription through the Tbx1 enhancer is consistent with a role for Foxa2 in linking Shh signaling to Tbx1 transcription in the endoderm.
The down-regulation of Foxc2 in Shh mutants is consistent with Foxc2 functioning as an intermediary between Shh signaling and Tbx1 regulation in the mesenchyme. The remaining Foxc1 expression in Shh mutants may be insufficient to activate Tbx1 fully, or Shh may be affecting Foxc1 activity through regulation of other cofactors. In either case, the evidence provided here of aortic arch defects in Shh mutants indicates that this molecular cascade is necessary for proper aortic arch patterning, although it remains to be determined whether Shh directly or indirectly controls Foxc2 expression. In addition, it remains possible that Shh regulates other pathways that also contribute to development of the aortic arch arteries.
Implications for the 22q11.2 deletion syndrome
Despite the relatively uniform microdeletions in patients with 22q11DS, a highly variable phenotype is commonly observed, possibly as a result of mutations or polymorphisms in functionally related genes. Because Shh and Fox proteins function in a common pathway with Tbx1, it will be interesting to determine if such upstream regulators serve as genetic modifiers of the 22q11DS phenotype. FOXC2 in particular may be such a modifier, as it directly regulates TBX1 and is responsible for a human syndrome that can be associated with cardiovascular defects, although typically in the setting of lymphedema and distichiasis (double-rowed eyebrows; Fang et al. 2000). The identification of a critical Tbx1 cis-regulatory element provides an opportunity to search for mutations of the Fox-binding element in patients with cardiac and craniofacial anomalies similar to those observed in patients with the 22q11 deletion. This will be particularly important as no mutations in the coding region of Tbx1 have been found in >200 patients with the DiGeorge phenotype. Mutation analyses of the cis- and trans-regulators of Tbx1 in patients with cranial and cardiac defects should ultimately reveal their significance in human disease.
Materials and methods
Generation of transgenic mice
We cloned a 5′ upstream genomic sequence of Tbx1 by screening a 129SV mouse genomic library of bacterial artificial clones (BAC; RPCI genomic BAC library from BACPAC Resources, Children's Hospital Oakland Research Institute). Different fragments of the Tbx1 upstream region were cloned into the hsp68lacZ reporter construct (Kothary et al. 1989). DNA for pronuclear injection was gel-purified and eluted using a QIAquick gel extraction kit (QIAGEN). Fertilized eggs from B6C3F1 female mice were collected for pronuclear injection. Injected eggs were implanted into ICR female mice, foster mothers were killed at E9.5, and F0 embryos were collected and stained for β-galactosidase (β-gal) activity as described previously (Yamagishi et al. 2000). Stable transgenic lines were obtained with a 12.8-kb fragment (construct 1) and a 1.1-kb fragment (construct 6), and F1 embryos were collected from E7.5 to E10.5 and also stained for β-gal activity. For histological analyses, embryos were dehydrated, embedded in paraffin, sectioned at 7 μm, rehydrated, and stained with Nuclear Fast Red (Moller and Moller 1994). Transgenes were detected by polymerase chain reaction (PCR) or Southern analysis for the lacZ gene in yolk sac DNA for embryos or tail DNA for 2-week-old pups. Primer sequences are available on request.
Mouse mutants
Mice with mutations in Shh (Chiang et al. 1996), Foxc1 (Kume et al. 1998), and Foxc2 (Winnier et al. 1997) were described previously. Shh mutant mice were used for intercrosses with Tbx1–lacZ or Tie2–lacZ (Schlaeger et al. 1997) transgenic mice.
Site-directed mutagenesis
The conserved Fox-binding site at −13,435 bp (TGTTTGT) was mutated to an NotI site in the context of the 1.1-kb fragment (−14,329/−13,230 bp). The 1.1-kb fragment was subcloned into plasmid pBluescript (Stratagene), and PCR-based mutagenesis on this plasmid was performed using pfu DNA polymerase and a QuikChange site-directed mutagenesis kit (Stratagene). The primers used were 5′-GGAGGAGCAGCCGCGGCCGCTT GCCAGATCTGTGC-3′ and 5′-GCACAGATCTGGCAAGCG GCCGCGGCTGCTCCTCC-3′. Restriction mapping and sequence analysis confirmed the presence of the mutation, and the insert was excised and cloned into the hsp68lacZ reporter plasmid.
Electrophoretic mobility shift assay (EMSA)
Oligonucleotides corresponding to the conserved Fox-binding site in the Tbx1 enhancer were synthesized (Integrated DNA Technologies) as follows: 5′-GGAGGAGCAGCCTGTTTGTT TTGCCAGATCTGTGC-3′, 5′-GCACAGATCTGGCAAAAC AAACAGGCTGCTCCTCC-3′. The primer pair for site-directed mutagenesis was used for mutant oligonucleotides. Oligonucleotide pairs were resuspended, boiled for 10 min, and annealed slowly, cooling to room temperature. Annealed oligonucleotides were radiolabeled with [α-32P]dCTP using a Klenow fragment of DNA polymerase and purified using Sephadex G-25 spin columns (Roche). Foxa2, Foxc1, and Foxc2 proteins were translated in vitro with the TNT T7-coupled reticulocyte lysate system (Promega), using full-length cDNA for Foxa2, Foxc1, and Foxc2 cloned into a pcDNA3.1 plasmid (Invitrogen) as DNA template. DNA-binding assays were performed as described previously (Scott et al. 1994). Unlabeled wild-type or mutant competitor oligonucleotide was added to reactions at the indicated concentrations. DNA–protein complexes were resolved on a 5% nondenaturing polyacrylamide gel in 0.5× TBE. Gels were exposed to Phosphor screen and read in PhosphorImager (Molecular Dynamics).
Transfection and luciferase assay
A nearly 200-bp DNA sequence (−13,546/−13,348 bp) containing the conserved Fox-binding site in the Tbx1 enhancer was amplified by PCR using primers 5′-CCCAAGCTTCCTGTGT GTCTGAGCTGGC-3′ and 5′-CGGGATCCTGCTGTCCACC CTGCAACCC-3′, and cloned into the pGL3 basic luciferase vector (Promega). Plasmid transfection was performed in 6-well plates using FuGENE 6 transfection reagent (Roche) according to the manufacturer's instructions. HeLa cells or COS-7 cells were transfected with 0.2 μg of luciferase reporter construct and various expression constructs, including 0.25 μg of pcDNA3.1 plasmids (as control), pcDNA3.1-Foxa2, pcDNA3.1-Foxc1, or pcDNA3.1-Foxc2. Then 0.1 μg of a RSV-lacZ expression construct was cotransfected to correct for transfection efficiency. Luciferase and lacZ activities in the cell extract were assayed 36–48 h after transfection, using the Luciferase Assay System (Promega) and Lucy2 luminometer (Rosys Anthos). All experiments were repeated at least twice, and representative results are shown.
In situ hybridization
35S-labeled antisense riboprobe was synthesized with T3 or SP6 RNA polymerase (MAXIscript in vitro transcription kit, Ambion) from Foxa2, Foxc1, Foxc2, or Tbx1 cDNA. Hybridization was performed on paraffin-embedded sections of E9.5–E10.5 wild type, Shh mutant, Foxc1 mutant, or Foxc2 mutant mouse embryos as described previously (Yamagishi et al. 2001). Sources of DNAs for making probes were as follows: Foxc1 and Foxc2 (Kume et al. 2001) and Tbx1 (Garg et al. 2001). Partial cDNA of Foxa2 was PCR-amplified using primer pair 5′-ATGC TGGGAGCCGTGAAGAT-3′ and 5′-CCAGTGACCAGAGC TAGCAG-3′ with full-length cDNA of Foxa2 [gift from K. Kaestner (University of Pennsylvania, Philadelphia)] as a template, and cloned into the pCRII-TOPO (Invitrogen) for generation of the probe.
Acknowledgments
We thank E.N. Olson for his support and critical reading of the manuscript, J.A. Richardson and members of the Molecular Pathology Core (J. Shelton, C. Pomajzl, J. Stark, D.L. Sutcliff) for assistance with histological analysis and section in situ hybridization, and S. Johnson for preparation of figures. We also thank C. Chiang for Shh mutant mice, T.N. Sato for Tie2–lacZ mice, and K. Kaestner and N. Miura for plasmids. This work was supported by a grant from the American Heart Association, Texas Affiliate (to H.Y.) and from the NHLBI/NIH, March of Dimes Birth Defects Foundation, Donald W. Reynolds Clinical Cardiovascular Center, and Smile Train Inc. (to D.S.). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1048903.
References
- Abu-Issa R, Smyth G, Smoak I, Yamamura KI, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF-3 β is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Bollag RJ, Siegfried Z, Cebra-Thomas JA, Garvey N, Davison EM, Silver LM. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T locus. Nat Genet. 1994;7:383–389. doi: 10.1038/ng0794-383. [DOI] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1–Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, Gorski JL, Seaver LH, Glover TW. Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema–distichiasis syndrome. Am J Hum Genet. 2000;67:1382–1388. doi: 10.1086/316915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Yamagishi C, Hu T, Kathiriya IS, Yamagishi H, Srivastava D. Tbx1, a DiGeorge syndrome candidate gene, is regulated by Sonic hedgehog during pharyngeal arch development. Dev Biol. 2001;235:62–73. doi: 10.1006/dbio.2001.0283. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Driscoll DA, Punnett HH, Sellinger B, Emanuel BS, Budarf ML. Characterization of 10p deletions suggests two nonoverlapping regions contribute to the DiGeorge syndrome phenotype. Am J Hum Genet. 1998;62:495–498. doi: 10.1086/301718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms JA, Kim CH, Hu D, Minkoff R, Thaller C, Eichele G. Sonic hedgehog participates in craniofacial morphogenesis and is down-regulated by teratogenic doses of retinoic acid. Dev Biol. 1997;187:25–35. doi: 10.1006/dbio.1997.8589. [DOI] [PubMed] [Google Scholar]
- Iida K, Koseki H, Kakinuma H, Kato N, Mizutani-Koseki Y, Ohuchi H, Yoshioka H, Noji S, Kawamura K, Kataoka Y, et al. Essential roles of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development. 1997;124:4627–4638. doi: 10.1242/dev.124.22.4627. [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes & Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68–lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93:985–996. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes & Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- Lindsay EA. Chromosomal microdeletions: Dissecting del22q11 syndrome. Nat Rev Genet. 2001;2:858–868. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Botta A, Jurecic V, Carattini-Rivera S, Cheah YC, Rosenblatt HM, Bradley A, Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, et al. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Martin G. Making a vertebrate limb: New players enter from the wings. Bioessays. 2001;23:865–868. doi: 10.1002/bies.1126. [DOI] [PubMed] [Google Scholar]
- McMahon AP. More surprises in the hedgehog signaling pathway. Cell. 2000;100:185–188. doi: 10.1016/s0092-8674(00)81555-x. [DOI] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Moller W, Moller G. Chemical dehydration for rapid paraffin embedding. Biotech Histochem. 1994;69:289–290. doi: 10.3109/10520299409106304. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, Silver LM. The T-box gene family. Bioessays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler T. Head and neck. In: Katz S, editor. Langman's medical embryology. 8th ed. Baltimore, MD: Lippincott, William & Wilkins; 2000. pp. 345–381. [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3β floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Scambler PJ. The 22q11 deletion syndromes. Hum Mol Genet. 2000;9:2421–2426. doi: 10.1093/hmg/9.16.2421. [DOI] [PubMed] [Google Scholar]
- Schilling TF. Genetic analysis of craniofacial development in the vertebrate embryo. Bioessays. 1997;19:459–468. doi: 10.1002/bies.950190605. [DOI] [PubMed] [Google Scholar]
- Schlaeger TM, Bartunkova S, Lawitts JA, Teichmann G, Risau W, Deutsch U, Sato TN. Uniform vascular-endothelial-cell-specific gene expression in both embryonic and adult transgenic mice. Proc Natl Acad Sci. 1997;94:3058–3063. doi: 10.1073/pnas.94.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott V, Clark AR, Docherty K. The gel retardation assay. In: Harwood AJ, editor. Methods in molecular biology, Vol. 31: Protocols for gene analysis. Totowa, NJ: Human Press; 1994. pp. 339–347. [DOI] [PubMed] [Google Scholar]
- Smith J. T-box genes: What they do and how they do it. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Morishima M, Taddei I, Lindsay EA, Baldini A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum Mol Genet. 2002a;11:915–922. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Taddei I, Morishima M, Meyers EN, Lindsay EA, Baldini A. A genetic link between Tbx1 and fibroblast growth factor signaling. Development. 2002b;129:4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr The winged-helix transcription factor HNF-3 β is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Winnier GE, Hargett L, Hogan BL. The winged helix transcription factor MFH1 is required for proliferation and patterning of paraxial mesoderm in the mouse embryo. Genes & Dev. 1997;11:926–940. doi: 10.1101/gad.11.7.926. [DOI] [PubMed] [Google Scholar]
- Winnier GE, Kume T, Deng K, Rogers R, Bundy J, Raines C, Walter MA, Hogan BL, Conway SJ. Roles for the winged helix transcription factors MF1 and MFH1 in cardiovascular development revealed by nonallelic noncomplementation of null alleles. Dev Biol. 1999;213:418–431. doi: 10.1006/dbio.1999.9382. [DOI] [PubMed] [Google Scholar]
- Yamagishi H. The 22q11.2 deletion syndrome. Keio J Med. 2002;51:77–88. doi: 10.2302/kjm.51.77. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Olson EN, Srivastava D. The basic helix–loop–helix transcription factor, dHAND, is required for vascular development. J Clin Invest. 2000;105:261–270. doi: 10.1172/JCI8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]